Running 'LAPS' Around nLD: Nuclear Lipid Droplet Form and Function
- PMID: 35178392
- PMCID: PMC8846306
- DOI: 10.3389/fcell.2022.837406
Running 'LAPS' Around nLD: Nuclear Lipid Droplet Form and Function
Abstract
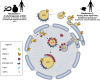
The nucleus harbours numerous protein subdomains and condensates that regulate chromatin organization, gene expression and genomic stress. A novel nuclear subdomain that is formed following exposure of cells to excess fatty acids is the nuclear lipid droplet (nLD), which is composed of a neutral lipid core surrounded by a phospholipid monolayer and associated regulatory and lipid biosynthetic enzymes. While structurally resembling cytoplasmic LDs, nLDs are formed by distinct but poorly understood mechanisms that involve the emergence of lipid droplets from the lumen of the nucleoplasmic reticulum and de novo lipid synthesis. Luminal lipid droplets that emerge into the nucleoplasm do so at regions of the inner nuclear membrane that become enriched in promyelocytic leukemia (PML) protein. The resulting nLDs that retain PML on their surface are termed lipid-associated PML structures (LAPS), and are distinct from canonical PML nuclear bodies (NB) as they lack key proteins and modifications associated with these NBs. PML is a key regulator of nuclear signaling events and PML NBs are sites of gene regulation and post-translational modification of transcription factors. Therefore, the subfraction of nLDs that form LAPS could regulate lipid stress responses through their recruitment and retention of the PML protein. Both nLDs and LAPS have lipid biosynthetic enzymes on their surface suggesting they are active sites for nuclear phospholipid and triacylglycerol synthesis as well as global lipid regulation. In this review we have summarized the current understanding of nLD and LAPS biogenesis in different cell types, their structure and composition relative to other PML-associated cellular structures, and their role in coordinating a nuclear response to cellular overload of fatty acids.
Keywords: CCTalpha; PML; fatty acid; lipin; nuclear lipid droplets; phosphatidylcholine.
Copyright © 2022 McPhee, Salsman, Foster, Thompson, Mathavarajah, Dellaire and Ridgway.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

