The role of SPAG1 in the assembly of axonemal dyneins in human airway epithelia
- PMID: 35178554
- PMCID: PMC8995097
- DOI: 10.1242/jcs.259512
The role of SPAG1 in the assembly of axonemal dyneins in human airway epithelia
Abstract
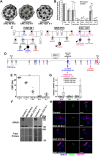
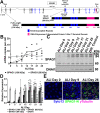
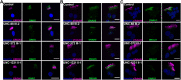
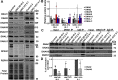
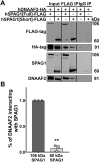
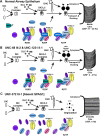
Mutations in SPAG1, a dynein axonemal assembly factor (DNAAF) that facilitates the assembly of dynein arms in the cytoplasm before their transport into the cilium, result in primary ciliary dyskinesia (PCD), a genetically heterogenous disorder characterized by chronic oto-sino-pulmonary disease, infertility and laterality defects. To further elucidate the role of SPAG1 in dynein assembly, we examined its expression, interactions and ciliary defects in control and PCD human airway epithelia. Immunoprecipitations showed that SPAG1 interacts with multiple DNAAFs, dynein chains and canonical components of the R2TP complex. Protein levels of dynein heavy chains (DHCs) and interactions between DHCs and dynein intermediate chains (DICs) were reduced in SPAG1 mutants. We also identified a previously uncharacterized 60 kDa SPAG1 isoform, through examination of PCD subjects with an atypical ultrastructural defect for SPAG1 variants, that can partially compensate for the absence of full-length SPAG1 to assemble a reduced number of outer dynein arms. In summary, our data show that SPAG1 is necessary for axonemal dynein arm assembly by scaffolding R2TP-like complexes composed of several DNAAFs that facilitate the folding and/or binding of the DHCs to the DIC complex.
Keywords: Dynein arm assembly; Motile cilia; Primary ciliary dyskinesia; R2TP complex; SPAG1.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

Similar articles
-
Mutations in SPAG1 cause primary ciliary dyskinesia associated with defective outer and inner dynein arms.Am J Hum Genet. 2013 Oct 3;93(4):711-20. doi: 10.1016/j.ajhg.2013.07.025. Epub 2013 Sep 19. Am J Hum Genet. 2013. PMID: 24055112 Free PMC article.
-
FBB18 participates in preassembly of almost all axonemal dyneins independent of R2TP complex.PLoS Genet. 2022 Aug 26;18(8):e1010374. doi: 10.1371/journal.pgen.1010374. eCollection 2022 Aug. PLoS Genet. 2022. PMID: 36026524 Free PMC article.
-
Role of the Novel Hsp90 Co-Chaperones in Dynein Arms' Preassembly.Int J Mol Sci. 2019 Dec 7;20(24):6174. doi: 10.3390/ijms20246174. Int J Mol Sci. 2019. PMID: 31817850 Free PMC article. Review.
-
Defects in the cytoplasmic assembly of axonemal dynein arms cause morphological abnormalities and dysmotility in sperm cells leading to male infertility.PLoS Genet. 2021 Feb 26;17(2):e1009306. doi: 10.1371/journal.pgen.1009306. eCollection 2021 Feb. PLoS Genet. 2021. PMID: 33635866 Free PMC article.
-
Ciliary Dyneins and Dynein Related Ciliopathies.Cells. 2021 Jul 25;10(8):1885. doi: 10.3390/cells10081885. Cells. 2021. PMID: 34440654 Free PMC article. Review.
Cited by
-
HYDIN Variants Are a Common Cause of Primary Ciliary Dyskinesia in French Canadians.Ann Am Thorac Soc. 2023 Jan;20(1):140-144. doi: 10.1513/AnnalsATS.202203-253RL. Ann Am Thorac Soc. 2023. PMID: 36112114 Free PMC article. No abstract available.
-
Promoter Deletion Leading to Allele Specific Expression in a Genetically Unsolved Case of Primary Ciliary Dyskinesia.Am J Med Genet A. 2025 Feb;197(2):e63880. doi: 10.1002/ajmg.a.63880. Epub 2024 Oct 4. Am J Med Genet A. 2025. PMID: 39364610
-
Strongly Truncated Dnaaf4 Plays a Conserved Role in Drosophila Ciliary Dynein Assembly as Part of an R2TP-Like Co-Chaperone Complex With Dnaaf6.Front Genet. 2022 Jul 6;13:943197. doi: 10.3389/fgene.2022.943197. eCollection 2022. Front Genet. 2022. PMID: 35873488 Free PMC article.
-
Formation and function of multiciliated cells.J Cell Biol. 2024 Jan 1;223(1):e202307150. doi: 10.1083/jcb.202307150. Epub 2023 Nov 30. J Cell Biol. 2024. PMID: 38032388 Free PMC article. Review.
References
-
- Austin-Tse, C., Halbritter, J., Zariwala, M. A., Gilberti, R. M., Gee, H. Y., Hellman, N., Pathak, N., Liu, Y., Panizzi, J. R., Patel-King, R. S.et al. (2013). Zebrafish ciliopathy screen plus human mutational analysis identifies C21orf59 and CCDC65 defects as causing primary ciliary dyskinesia. Am. J. Hum. Genet. 93, 672-686. 10.1016/j.ajhg.2013.08.015 - DOI - PMC - PubMed
-
- Benbahouche, N. E. H., Iliopoulos, I., Török, I., Marhold, J., Henri, J., Kajava, A. V., Farkaš, R., Kempf, T., Schnölzer, M., Meyer, P.et al. (2014). Drosophila Spag is the homolog of RNA polymerase II-associated protein 3 (RPAP3) and recruits the heat shock proteins 70 and 90 (Hsp70 and Hsp90) during the assembly of cellular machineries. J. Biol. Chem. 289, 6236-6247. 10.1074/jbc.M113.499608 - DOI - PMC - PubMed
-
- Biermann, K., Heukamp, L. C., Steger, K., Zhou, H., Franke, F. E., Sonnack, V., Brehm, R., Berg, J., Bastian, P. J. and Müller, S. C. (2007). Genome-wide expression profiling reveals new insights into pathogenesis and progression of testicular germ cell tumors. Cancer Genomics Proteomics 4, 359-367. - PubMed
-
- Bio-Rad. (2018). Droplet DigitalTM PCR Droplet DigitalTM PCR Applications Guide. Bulletin 6407, 1-145.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

