The RD-Connect Genome-Phenome Analysis Platform: Accelerating diagnosis, research, and gene discovery for rare diseases
- PMID: 35178824
- PMCID: PMC9324157
- DOI: 10.1002/humu.24353
The RD-Connect Genome-Phenome Analysis Platform: Accelerating diagnosis, research, and gene discovery for rare diseases
Abstract
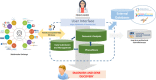
Rare disease patients are more likely to receive a rapid molecular diagnosis nowadays thanks to the wide adoption of next-generation sequencing. However, many cases remain undiagnosed even after exome or genome analysis, because the methods used missed the molecular cause in a known gene, or a novel causative gene could not be identified and/or confirmed. To address these challenges, the RD-Connect Genome-Phenome Analysis Platform (GPAP) facilitates the collation, discovery, sharing, and analysis of standardized genome-phenome data within a collaborative environment. Authorized clinicians and researchers submit pseudonymised phenotypic profiles encoded using the Human Phenotype Ontology, and raw genomic data which is processed through a standardized pipeline. After an optional embargo period, the data are shared with other platform users, with the objective that similar cases in the system and queries from peers may help diagnose the case. Additionally, the platform enables bidirectional discovery of similar cases in other databases from the Matchmaker Exchange network. To facilitate genome-phenome analysis and interpretation by clinical researchers, the RD-Connect GPAP provides a powerful user-friendly interface and leverages tens of information sources. As a result, the resource has already helped diagnose hundreds of rare disease patients and discover new disease causing genes.
Keywords: NGS; data sharing; data standardization; diagnostics; genome analysis; patient matchmaking; rare diseases.
© 2022 The Authors. Human Mutation published by Wiley Periodicals LLC.
Conflict of interest statement
Orion Buske is the CEO of PhenoTips. Jules Jacobsen is a consultant to Congenica. All other authors declare no conflict of interest.
Figures
References
-
- Austin, C. P. , Cutillo, C. M. , Lau, L. , Jonker, A. H. , Rath, A. , Julkowska, D. , Thomson, D. , Terry, S. F. , de Montleau, B. , Ardigò, D. , Hivert, V. , Boycott, K. M. , Baynam, G. , Kaufmann, P. , Taruscio, D. , Lochmüller, H. , Suematsu, M. , Incerti, C. , Draghia‐Akli, R. , … International Rare Diseases Research Consortium (2018). Future of rare diseases research 2017–2027: An IRDiRC perspective. Clinical and Translational Science, 11, 21–27. 10.1111/cts.12500 - DOI - PMC - PubMed
-
- Boycott, K. M. , Rath, A. , Chong, J. X. , Hartley, T. , Alkuraya, F. S. , Baynam, G. , Brookes, A. J. , Brudno, M. , Carracedo, A. , den Dunnen, J. T. , Dyke, S. , Estivill, X. , Goldblatt, J. , Gonthier, C. , Groft, S. C. , Gut, I. , Hamosh, A. , Hieter, P. , Höhn, S. , … Lochmüller, H. (2017). International cooperation to enable the diagnosis of all rare genetic diseases. American Journal of Human Genetics, 100, 695–705. 10.1016/j.ajhg.2017.04.003 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

