Aberrant translation regulated by METTL1/WDR4-mediated tRNA N7-methylguanosine modification drives head and neck squamous cell carcinoma progression
- PMID: 35179319
- PMCID: PMC8923133
- DOI: 10.1002/cac2.12273
Aberrant translation regulated by METTL1/WDR4-mediated tRNA N7-methylguanosine modification drives head and neck squamous cell carcinoma progression
Abstract
Background: Cancer cells selectively promote the translation of oncogenic transcripts to stimulate cancer progression. Although growing evidence has revealed that tRNA modifications and related genes participate in this process, their roles in head and neck squamous cell carcinoma (HNSCC) remain largely uncharacterized. Here, we sought to investigate the function and mechanisms of the transfer RNA (tRNA) N7-methylguanosine (m7 G) modification in regulating the occurrence and development of HNSCC.
Methods: Cell lost-of-function and gain-of-function assays, xenograft models, conditional knockout and knockin mouse models were used to study the physiological functions of tRNA m7 G modification in HNSCC tumorigenesis. tRNA modification and expression profiling, mRNA translation profiling and rescue assays were performed to uncover the underlying molecular mechanisms. Single-cell RNA sequencing (scRNA-seq) was conducted to explore the tumor microenvironment changes.
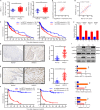
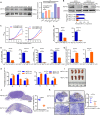
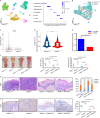
Results: The tRNA m7 G methyltransferase complex components Methyltransferase-like 1 (METTL1)/WD repeat domain 4 (WDR4) were upregulated in HNSCC and associated with a poor prognosis. Functionally, METTL1/WDR4 promoted HNSCC progression and metastasis in cell-based and transgenic mouse models. Mechanistically, ablation of METTL1 reduced the m7 G levels of 16 tRNAs, inhibiting the translation of a subset of oncogenic transcripts, including genes related to the phosphatidylinositol-3-kinase/protein kinase B/mammalian target of rapamycin (PI3K/AKT/mTOR) signaling pathway. In addition, chemical modulators of the PI3K/Akt/mTOR signaling pathway reversed the effects of Mettl1 in mouse HNSCC. Furthermore, scRNA-seq results revealed that Mettl1 knockout in mouse tumor cells altered the immune landscape and cell-cell interaction between the tumor and stromal compartment.
Conclusions: The tRNA m7 G methyltransferase METTL1 was found to promote the development and malignancy of HNSCC through regulating global mRNA translation, including the PI3K/AKT/mTOR signaling pathway, and found to alter immune landscape. METTL1 could be a promising treatment target for HNSCC patients.
Keywords: METTL1; PI3K/AKT/mTOR signaling; WDR4; head and neck squamous cell carcinoma; m7G modification; metastasis; microenvironment; scRNA-seq; tRNA.
© 2022 The Authors. Cancer Communications published by John Wiley & Sons Australia, Ltd. on behalf of Sun Yat-sen University Cancer Center.
Conflict of interest statement
The authors have declared that no conflict of interest exists.
Figures




References
-
- Santos M, Fidalgo A, Varanda AS, Oliveira C, Santos MAS. tRNA Deregulation and Its Consequences in Cancer. Trends Mol Med. 2019;25(10):853‐65. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

