Towards the prediction of non-peptidic epitopes
- PMID: 35180214
- PMCID: PMC8893639
- DOI: 10.1371/journal.pcbi.1009151
Towards the prediction of non-peptidic epitopes
Abstract
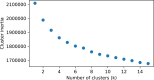
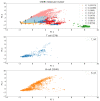
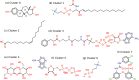
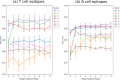
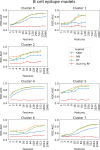
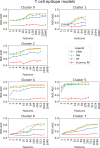
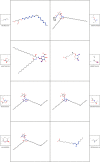
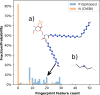
In-silico methods for the prediction of epitopes can support and improve workflows for vaccine design, antibody production, and disease therapy. So far, the scope of B cell and T cell epitope prediction has been directed exclusively towards peptidic antigens. Nevertheless, various non-peptidic molecular classes can be recognized by immune cells. These compounds have not been systematically studied yet, and prediction approaches are lacking. The ability to predict the epitope activity of non-peptidic compounds could have vast implications; for example, for immunogenic risk assessment of the vast number of drugs and other xenobiotics. Here we present the first general attempt to predict the epitope activity of non-peptidic compounds using the Immune Epitope Database (IEDB) as a source for positive samples. The molecules stored in the Chemical Entities of Biological Interest (ChEBI) database were chosen as background samples. The molecules were clustered into eight homogeneous molecular groups, and classifiers were built for each cluster with the aim of separating the epitopes from the background. Different molecular feature encoding schemes and machine learning models were compared against each other. For those models where a high performance could be achieved based on simple decision rules, the molecular features were then further investigated. Additionally, the findings were used to build a web server that allows for the immunogenic investigation of non-peptidic molecules (http://tools-staging.iedb.org/np_epitope_predictor). The prediction quality was tested with samples from independent evaluation datasets, and the implemented method received noteworthy Receiver Operating Characteristic-Area Under Curve (ROC-AUC) values, ranging from 0.69-0.96 depending on the molecule cluster.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

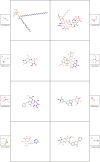
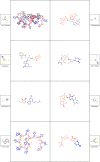
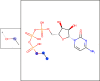
References
-
- Chow SN, Chen KW, Su SL, Tung J, Lee CY. Generation and epitope analysis of thyroid stimulating hormone-specific monoclonal antibodies for enzyme immunoassays. Biotechnol Appl Biochem. 1988;10: 137–142. - PubMed
-
- Paraf A, Peltre G. Immunoassays in Food and Agriculture. Springer Netherlands; 1991.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

