Unresolved stalled ribosome complexes restrict cell-cycle progression after genotoxic stress
- PMID: 35180429
- PMCID: PMC9098122
- DOI: 10.1016/j.molcel.2022.01.019
Unresolved stalled ribosome complexes restrict cell-cycle progression after genotoxic stress
Abstract
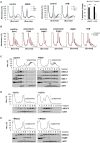
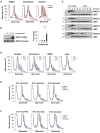
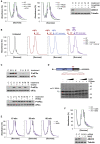
During the translation surveillance mechanism known as ribosome-associated quality control, the ASC-1 complex (ASCC) disassembles ribosomes stalled on the mRNA. Here, we show that there are two distinct classes of stalled ribosome. Ribosomes stalled by translation elongation inhibitors or methylated mRNA are short lived in human cells because they are split by the ASCC. In contrast, although ultraviolet light and 4-nitroquinoline 1-oxide induce ribosome stalling by damaging mRNA, and the ASCC is recruited to these stalled ribosomes, we found that they are refractory to the ASCC. Consequently, unresolved UV- and 4NQO-stalled ribosomes persist in human cells. We show that ribosome stalling activates cell-cycle arrest, partly through ZAK-p38MAPK signaling, and that this cell-cycle delay is prolonged when the ASCC cannot resolve stalled ribosomes. Thus, we propose that the sensitivity of stalled ribosomes to the ASCC influences the kinetics of stall resolution, which in turn controls the adaptive stress response.
Keywords: ASC-1 complex; RNA damage; RNA-binding protein; cell-cycle arrest; ribosome stalling; ribosome-associated quality control; ultraviolet light.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.E.W. is a member of the Molecular Cell advisory board.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

