Obesity-induced senescent macrophages activate a fibrotic transcriptional program in adipocyte progenitors
- PMID: 35181634
- PMCID: PMC8860101
- DOI: 10.26508/lsa.202101286
Obesity-induced senescent macrophages activate a fibrotic transcriptional program in adipocyte progenitors
Abstract
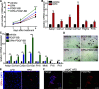
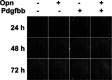
Adipose tissue fibrosis is regulated by the chronic and progressive metabolic imbalance caused by differences in caloric intake and energy expenditure. By exploring the cellular heterogeneity within fibrotic adipose tissue, we demonstrate that early adipocyte progenitor cells expressing both platelet-derived growth factor receptor (PDGFR) α and β are the major contributors to extracellular matrix deposition. We show that the fibrotic program is promoted by senescent macrophages. These macrophages were enriched in the fibrotic stroma and exhibit a distinct expression profile. Furthermore, we demonstrate that these cells display a blunted phagocytotic capacity and acquire a senescence-associated secretory phenotype. Finally, we determined that osteopontin, which was expressed by senescent macrophages in the fibrotic environment promoted progenitor cell proliferation, fibrotic gene expression, and inhibited adipogenesis. Our work reveals that obesity promotes macrophage senescence and provides a conceptual framework for the discovery of rational therapeutic targets for metabolic and inflammatory disease associated with obesity.
© 2022 Rabhi et al.
Figures








References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
