Biocatalysed synthesis planning using data-driven learning
- PMID: 35181654
- PMCID: PMC8857209
- DOI: 10.1038/s41467-022-28536-w
Biocatalysed synthesis planning using data-driven learning
Abstract
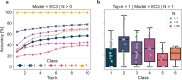
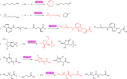
Enzyme catalysts are an integral part of green chemistry strategies towards a more sustainable and resource-efficient chemical synthesis. However, the use of biocatalysed reactions in retrosynthetic planning clashes with the difficulties in predicting the enzymatic activity on unreported substrates and enzyme-specific stereo- and regioselectivity. As of now, only rule-based systems support retrosynthetic planning using biocatalysis, while initial data-driven approaches are limited to forward predictions. Here, we extend the data-driven forward reaction as well as retrosynthetic pathway prediction models based on the Molecular Transformer architecture to biocatalysis. The enzymatic knowledge is learned from an extensive data set of publicly available biochemical reactions with the aid of a new class token scheme based on the enzyme commission classification number, which captures catalysis patterns among different enzymes belonging to the same hierarchy. The forward reaction prediction model (top-1 accuracy of 49.6%), the retrosynthetic pathway (top-1 single-step round-trip accuracy of 39.6%) and the curated data set are made publicly available to facilitate the adoption of enzymatic catalysis in the design of greener chemistry processes.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







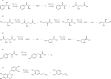
References
-
- Antony, T. Malthus foiled again and again. Nature418, 668–670 (2002). - PubMed
-
- Matlin, S. A. & Abegaz, B. M. In The Chemical Element: Chemistry’s Contribution to Our Global Future. (eds García-Martínez, J., Serrano-Torregrosa, E.) (Wiley-VCH, 2011).
-
- Zimmerman, J. B., Anastas, P. T., Erythropel, H. C. & Walter, L. Designing for a green chemistry future. Science367, 397–400 (2020). - PubMed
-
- Stanislav, M., Zbynek, P. & Jiri, D. Machine learning in enzyme engineering. ACS Catal.10, 1210–1223 (2020).
-
- Homaei, A. A., Reyhaneh, S., Fabio, V. & Roberto, S. Enzyme immobilization: an update. J. Chem. Biol.https://link.springer.com/article/10.1007/s12154-013-0102-9 (2013). - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

