Discovery of actinomycin L, a new member of the actinomycin family of antibiotics
- PMID: 35181725
- PMCID: PMC8857259
- DOI: 10.1038/s41598-022-06736-0
Discovery of actinomycin L, a new member of the actinomycin family of antibiotics
Abstract
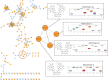
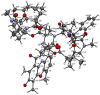
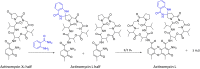
Streptomycetes are major producers of bioactive natural products, including the majority of the naturally produced antibiotics. While much of the low-hanging fruit has been discovered, it is predicted that less than 5% of the chemical space of natural products has been mined. Here, we describe the discovery of the novel actinomycins L1 and L2 produced by Streptomyces sp. MBT27, via application of metabolic analysis and molecular networking. Actinomycins L1 and L2 are diastereomers, and the structure of actinomycin L2 was resolved using NMR and single crystal X-ray crystallography. Actinomycin L is formed via spirolinkage of anthranilamide to the 4-oxoproline moiety of actinomycin X2, prior to the condensation of the actinomycin halves. Such a structural feature has not previously been identified in naturally occurring actinomycins. Adding anthranilamide to cultures of the actinomycin X2 producer Streptomyces antibioticus, which has the same biosynthetic gene cluster as Streptomyces sp. MBT27, resulted in the production of actinomycin L. This supports a biosynthetic pathway whereby actinomycin L is produced from two distinct metabolic routes, namely those for actinomycin X2 and for anthranilamide. Actinomycins L1 and L2 showed significant antimicrobial activity against Gram-positive bacteria. Our work shows how new molecules can still be identified even in the oldest of natural product families.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

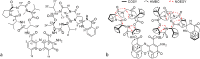


References
-
- Wright GD. Solving the antibiotic crisis. ACS Infect. Dis. 2015;1:80–84. - PubMed
-
- Bérdy J. Bioactive microbial metabolites. J. Antibiot. (Tokyo) 2005;58:1–26. - PubMed
-
- Bentley SD, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

