KANADI promotes thallus differentiation and FR-induced gametangiophore formation in the liverwort Marchantia
- PMID: 35181887
- PMCID: PMC9311212
- DOI: 10.1111/nph.18046
KANADI promotes thallus differentiation and FR-induced gametangiophore formation in the liverwort Marchantia
Abstract
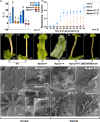
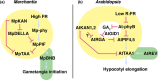
In angiosperms, KANADI transcription factors have roles in the sporophyte generation regulating tissue polarity, organogenesis and shade avoidance responses, but are not required during the gametophyte generation. Whether these roles are conserved in the gametophyte-dominant bryophyte lineages is unknown, which we examined by characterising the sole KANADI ortholog, MpKAN, in the liverwort Marchantia polymorpha. In contrast to angiosperm orthologs, MpKAN functions in the gametophyte generation in Marchantia, where it regulates apical branching and tissue differentiation, but does not influence tissue polarity in either generation. MpKAN can partially rescue the sporophyte polarity defects of kanadi mutants in Arabidopsis, indicating that MpKAN has conserved biochemical activity to its angiosperm counterparts. Mpkan loss-of-function plants display defects in far-red (FR) light responses. Mpkan plants have reduced FR-induced growth tropisms, have a delayed transition to sexual reproduction and fail to correctly form gametangiophores. Our results indicate that MpKAN is a modulator of FR responses, which may reflect a conserved role for KANADI across land plants. Under FR, MpKAN negatively regulates MpDELLA expression, suggesting that MpKAN and MpDELLA act in a pathway regulating FR responses, placing MpKAN in a gene regulatory network exhibiting similarities with those of angiosperms.
Keywords: Marchantia; KANADI; far-red (FR) responses; gametangiophores; land plant evolution; transcription factor evolution.
© 2022 The Authors. New Phytologist © 2022 New Phytologist Foundation.
Figures




References
-
- Albert NW, Thrimawithana AH, McGhie TK, Clayton WA, Deroles SC, Schwinn KE, Bowman JL, Jordan BR, Davies KM. 2018. Genetic analysis of the liverwort Marchantia polymorpha reveals that R2R3MYB activation of flavonoid production in response to abiotic stress is an ancient character in land plants. New Phytologist 218: 554–566. - PubMed
-
- Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F et al. 2017. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171: 287–304 e15. - PubMed
-
- Brandt R, Salla‐Martret M, Bou‐Torrent J, Musielak T, Stahl M, Lanz C, Ott F, Schmid M, Greb T, Schwarz M et al. 2012. Genome‐wide binding‐site analysis of REVOLUTA reveals a link between leaf patterning and light‐mediated growth responses. The Plant Journal 72: 31–42. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

