Functionally prioritised whole-genome sequence variants improve the accuracy of genomic prediction for heat tolerance
- PMID: 35183109
- PMCID: PMC8858496
- DOI: 10.1186/s12711-022-00708-8
Functionally prioritised whole-genome sequence variants improve the accuracy of genomic prediction for heat tolerance
Abstract
Background: Heat tolerance is a trait of economic importance in the context of warm climates and the effects of global warming on livestock production, reproduction, health, and well-being. This study investigated the improvement in prediction accuracy for heat tolerance when selected sets of sequence variants from a large genome-wide association study (GWAS) were combined with a standard 50k single nucleotide polymorphism (SNP) panel used by the dairy industry.
Methods: Over 40,000 dairy cattle with genotype and phenotype data were analysed. The phenotypes used to measure an individual's heat tolerance were defined as the rate of decline in milk production traits with rising temperature and humidity. We used Holstein and Jersey cows to select sequence variants linked to heat tolerance. The prioritised sequence variants were the most significant SNPs passing a GWAS p-value threshold selected based on sliding 100-kb windows along each chromosome. We used a bull reference set to develop the genomic prediction equations, which were then validated in an independent set of Holstein, Jersey, and crossbred cows. Prediction analyses were performed using the BayesR, BayesRC, and GBLUP methods.
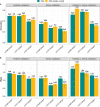
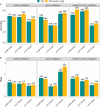
Results: The accuracy of genomic prediction for heat tolerance improved by up to 0.07, 0.05, and 0.10 units in Holstein, Jersey, and crossbred cows, respectively, when sets of selected sequence markers from Holstein cows were added to the 50k SNP panel. However, in some scenarios, the prediction accuracy decreased unexpectedly with the largest drop of - 0.10 units for the heat tolerance fat yield trait observed in Jersey cows when 50k plus pre-selected SNPs from Holstein cows were used. Using pre-selected SNPs discovered on a combined set of Holstein and Jersey cows generally improved the accuracy, especially in the Jersey validation. In addition, combining Holstein and Jersey bulls in the reference set generally improved prediction accuracy in most scenarios compared to using only Holstein bulls as the reference set.
Conclusions: Informative sequence markers can be prioritised to improve the genomic prediction of heat tolerance in different breeds. In addition to providing biological insight, these variants could also have a direct application for developing customized SNP arrays or can be used via imputation in current industry SNP panels.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Polsky L, von Keyserlingk MA. Invited review: Effects of heat stress on dairy cattle welfare. J Dairy Sci. 2017;100:8645–8657. - PubMed
-
- St-Pierre N, Cobanov B, Schnitkey G. Economic losses from heat stress by US livestock industries. J Dairy Sci. 2003;86:E52–77.
-
- Carabaño MJ, Ramón M, Díaz C, Molina A, Pérez-Guzmán MD, Serradilla JM. Breeding and genetics symposium: breeding for resilience to heat stress effects in dairy ruminants. A comprehensive review. J Anim Sci. 2017;95:1813–1826. - PubMed
-
- Ravagnolo O, Misztal I, Hoogenboom G. Genetic component of heat stress in dairy cattle, development of heat index function. J Dairy Sci. 2000;83:2120–2125. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

