Identification of a WRKY transcriptional activator from Camellia sinensis that regulates methylated EGCG biosynthesis
- PMID: 35184160
- PMCID: PMC9071374
- DOI: 10.1093/hr/uhac024
Identification of a WRKY transcriptional activator from Camellia sinensis that regulates methylated EGCG biosynthesis
Abstract
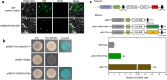
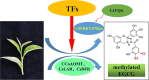
Naturally occurring methylated catechins, especially methylated EGCG in tea leaves are known to have many health benefits. Although the genes involved in methylated EGCG biosynthesis have been studied extensively, the transcriptional factors controlling methylated EGCG biosynthesis are still poorly understood. In the present study, a WRKY domain-containing protein termed CsWRKY57like was identified, which belongs to group IIc of the WRKY family, and contains one conserved WRKY motif. CsWRKY57like was found to localize in the nucleus, function as a transcriptional activator, and its expression positively correlated with methylated EGCG level. In addition, CsWRKY57like activated the transcription of three genes related to methylated EGCG biosynthesis, including CCoAOMT, CsLAR, and CsDFR by specifically interacting with their promoters via binding to the cis-acting element (C/T)TGAC(T/C). Further assays revealed that CsWRKY57like physically interacts with CsVQ4, and participates in the metabolic regulation of O-methylated catechin biosynthesis. Collectively, we conclude that CsWRKY57like may positively impact the biosynthesis of methylated EGCG in the tea plant, which comprehensively enriches the regulatory network of WRKY TFs associated with methylated EGCG and provide a potential strategy for the breeding of specific tea plant cultivars with high methylated EGCG .
Keywords: Camellia sinensis; Methylated EGCG; WRKY; transcriptional regulation.
© The Author(s) 2022. Published by Oxford University Press. All rights reserved.
Figures





References
-
- Zhu JY, Xu Q, Zhao Set al. . Comprehensive co-expression analysis provides novel insights into temporal variation of flavonoids in fresh leaves of the tea plant (Camellia sinensis). Plant Sci. 2020;290:110306. - PubMed
-
- Donlao N, Ogawa Y. The influence of processing conditions on catechin, caffeine and chlorophyll contents of green tea (Camelia sinensis) leaves and infusions. LWT. 2019;116:108567.
-
- Ng KW, Cao Z-J, Chen H-Bet al. . Oolong tea: a critical review of processing methods, chemical composition, health effects, and risk. Crit Rev Food Sci Nutr. 2018;58:2957–80. - PubMed
LinkOut - more resources
Full Text Sources

