Genetic architecture and genomic predictive ability of apple quantitative traits across environments
- PMID: 35184165
- PMCID: PMC8976694
- DOI: 10.1093/hr/uhac028
Genetic architecture and genomic predictive ability of apple quantitative traits across environments
Erratum in
-
Correction to: Genetic architecture and genomic predictive ability of apple quantitative traits across environments.Hortic Res. 2022 Feb 19;9:uhac273. doi: 10.1093/hr/uhac273. eCollection 2022. Hortic Res. 2022. PMID: 36518184 Free PMC article.
Abstract
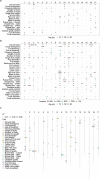
Implementation of genomic tools is desirable to increase the efficiency of apple breeding. Recently, the multi-environment apple reference population (apple REFPOP) proved useful for rediscovering loci, estimating genomic predictive ability, and studying genotype by environment interactions (G × E). So far, only two phenological traits were investigated using the apple REFPOP, although the population may be valuable when dissecting genetic architecture and reporting predictive abilities for additional key traits in apple breeding. Here we show contrasting genetic architecture and genomic predictive abilities for 30 quantitative traits across up to six European locations using the apple REFPOP. A total of 59 stable and 277 location-specific associations were found using GWAS, 69.2% of which are novel when compared with 41 reviewed publications. Average genomic predictive abilities of 0.18-0.88 were estimated using main-effect univariate, main-effect multivariate, multi-environment univariate, and multi-environment multivariate models. The G × E accounted for up to 24% of the phenotypic variability. This most comprehensive genomic study in apple in terms of trait-environment combinations provided knowledge of trait biology and prediction models that can be readily applied for marker-assisted or genomic selection, thus facilitating increased breeding efficiency.
© The Author(s) 2022. Published by Oxford University Press. All rights reserved.
Figures






References
-
- FAOSTAT (Food and Agriculture Organization of the United Nations, 2019).
-
- Way RD, Aldwinckle HS, Lamb RCet al. . Apples (Malus). Acta Hortic. 1991;3–46. 10.17660/ActaHortic.1991.290.1. - DOI
LinkOut - more resources
Full Text Sources

