Esculin hydrolysis negative and TcdA-only producing strains of Clostridium (Clostridioides) difficile from the environment in Western Australia
- PMID: 35184359
- PMCID: PMC9544920
- DOI: 10.1111/jam.15500
Esculin hydrolysis negative and TcdA-only producing strains of Clostridium (Clostridioides) difficile from the environment in Western Australia
Abstract
Background and aims: Clostridium (Clostridiodes) difficile clade 3 ribotype (RT) 023 strains that fail to produce black colonies on bioMérieux ChromID agar have been reported, as well as variant strains of C. difficile that produce only toxin A. We have recently isolated strains of C. difficile from the environment in Western Australia (WA) with similar characteristics. The objective of this study was to characterize these strains. It was hypothesized that a putative β-glucosidase gene was lacking in these strains of C. difficile, including RT 023, leading to white colonies.
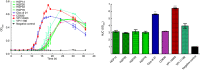
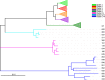
Methods and results: A total of 17 environmental isolates of C. difficile from garden soil and compost, and gardening shoe soles in Perth, WA, failed to produce black colonies on ChromID agar. MALDI-TOF MS analysis confirmed these strains as C. difficile. Four strains contained only a tcdA gene (A+ B- CDT- ) by PCR and were a novel RT (QX 597). All isolates were susceptible to all antimicrobials tested except one with low-level resistance to clindamycin (MIC = 8 mg/L). The four tcdA-positive strains were motile. All isolates contained neither bgl locus but only bgl K or a putative β-glucosidase gene by PCR. Whole-genome sequencing showed the 17 strains belonged to novel multi-locus sequence types 632, 848, 849, 850, 851, 852 and 853, part of the evolutionarily divergent clade C-III. Four isolates carried a full-length tcdA but not tcdB nor binary toxin genes.
Conclusions: ChromID C. difficile agar is used for the specific detection of C. difficile in the samples. To date, all strains except RT 023 strains from clinical samples hydrolyse esculin. This is the first report to provide insights into the identification of esculin hydrolysis negative and TcdA-only producing (A+ B- CDT- ) strains of C. difficile from environmental samples.
Significance and impact of the study: White colonies of C. difficile from environmental samples could be overlooked when using ChromID C. difficile agar, leading to false-negative results, however, whether these strains are truly pathogenic remains to be proven.
Keywords: Clostridium (Clostridioides) difficile; clade-III; environment; esculin hydrolysis negative; putative β glucosidase gene; toxin A variant; whole-genome sequencing.
© 2022 The Authors. Journal of Applied Microbiology published by John Wiley & Sons Ltd on behalf of Society for Applied Microbiology.
Conflict of interest statement
No conflict of interest was declared.
Figures


References
-
- Anderson, D.J. , Rojas, L.F. , Watson, S. , Knelson, L.P. , Pruitt, S. , Lewis, S.S. et al. (2017) Identification of novel risk factors for community‐acquired Clostridium difficile infection using spatial statistics and geographic information system analyses. PLoS ONE, 12, e0176285. 10.1371/journal.pone.0176285 - DOI - PMC - PubMed
-
- Androga, G.O. , Knight, D.R. , Hutton, M.L. , Mileto, S.J. , James, M.L. , Evans, C. et al. (2019) In silico, in vitro and in vivo analysis of putative virulence factors identified in large clostridial toxin‐negative, binary toxin‐producing Clostridium difficile strains. Anaerobe, 60, 102083. - PubMed
-
- Clinical and Laboratory Standards Institute (CLSI) . (2007) Methods for antimicrobial susceptibility testing of anaerobic bacteria. Approved Standard—Seventh Edition. CLSI document M11‐A7 [ISBN 1‐56238626‐3]. Wayne: Clinical and Laboratory Standards Institute.
-
- Clinical and Laboratory Standards Institute (CLSI) . (2013) Performance standards for antimicrobial susceptibility testing; Twenty‐Third Informational Supplement. CLSI document M100‐S23. Wayne: Clinical and Laboratory Standards Institute.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

