CHML is an NRF2 target gene that regulates mTOR function
- PMID: 35184380
- PMCID: PMC9019883
- DOI: 10.1002/1878-0261.13194
CHML is an NRF2 target gene that regulates mTOR function
Abstract
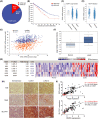
The transcription factor nuclear factor erythroid 2-related factor 2 (NRF2) is often highly expressed in non-small cell lung cancer (NSCLC). Through its target genes, NRF2 enhances cancer progression and chemo/radioresistance, leading to a poorer prognosis in patients with high NRF2 expression. In this study, we identified CHM-like Rab escort protein (CHML; encoding Rep2) as an NRF2 target gene with an antioxidant response element (ARE) in its promoter region (-1622 to -1612). Analysis of patient data curated by The Cancer Genome Atlas (TCGA) and Oncomine databases revealed that CHML mRNA expression was elevated in lung adenocarcinoma (LUAD) patient tumor tissues and correlated with decreased patient survival. Immunohistochemistry (IHC) analysis of normal versus lung cancer patient tissues revealed that Rep2 protein levels were higher in lung tumors compared with normal tissue, which also correlated with increased levels of NRF2. Importantly, siRNA-mediated knockdown of CHML/Rep2 in A549 NSCLC cells decreased their ability to proliferate. Mechanistically, Rep2 mediates mTOR function, as loss of Rep2 inhibited, whereas overexpression enhanced, mTOR translocation and activation at the lysosome. Our findings identify a novel NRF2-Rep2-dependent regulation of mTOR function.
Keywords: CHML/REP2; NRF2; NSCLC; mTOR.
© 2022 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

