Genotypic Variation of Nitrogen Use Efficiency and Amino Acid Metabolism in Barley
- PMID: 35185958
- PMCID: PMC8854266
- DOI: 10.3389/fpls.2021.807798
Genotypic Variation of Nitrogen Use Efficiency and Amino Acid Metabolism in Barley
Erratum in
-
Corrigendum: Genotypic Variation of Nitrogen Use Efficiency and Amino Acid Metabolism in Barley.Front Plant Sci. 2022 Apr 5;13:893540. doi: 10.3389/fpls.2022.893540. eCollection 2022. Front Plant Sci. 2022. PMID: 35449892 Free PMC article.
Abstract
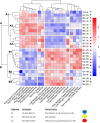
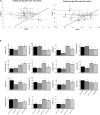
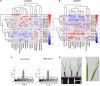
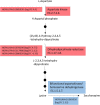
Owing to the large genetic diversity of barley and its resilience under harsh environments, this crop is of great value for agroecological transition and the need for reduction of nitrogen (N) fertilizers inputs. In the present work, we investigated the diversity of a North African barley genotype collection in terms of growth under limiting N (LN) or ample N (HN) supply and in terms of physiological traits including amino acid content in young seedlings. We identified a Moroccan variety, Laanaceur, accumulating five times more lysine in its leaves than the others under both N nutritional regimes. Physiological characterization of the barley collection showed the genetic diversity of barley adaptation strategies to LN and highlighted a genotype x environment interaction. In all genotypes, N limitation resulted in global biomass reduction, an increase in C concentration, and a higher resource allocation to the roots, indicating that this organ undergoes important adaptive metabolic activity. The most important diversity concerned leaf nitrogen use efficiency (LNUE), root nitrogen use efficiency (RNUE), root nitrogen uptake efficiency (RNUpE), and leaf nitrogen uptake efficiency (LNUpE). Using LNUE as a target trait reflecting barley capacity to deal with N limitation, this trait was positively correlated with plant nitrogen uptake efficiency (PNUpE) and RNUpE. Based on the LNUE trait, we determined three classes showing high, moderate, or low tolerance to N limitation. The transcriptomic approach showed that signaling, ionic transport, immunity, and stress response were the major functions affected by N supply. A candidate gene encoding the HvNRT2.10 transporter was commonly up-regulated under LN in the three barley genotypes investigated. Genes encoding key enzymes required for lysine biosynthesis in plants, dihydrodipicolinate synthase (DHPS) and the catabolic enzyme, the bifunctional Lys-ketoglutarate reductase/saccharopine dehydrogenase are up-regulated in Laanaceur and likely account for a hyperaccumulation of lysine in this genotype. Our work provides key physiological markers of North African barley response to low N availability in the early developmental stages.
Keywords: NUE (nitrogen use efficiency); barley; crop/stress physiology; lysine (amino acids); natural variability.
Copyright © 2022 Decouard, Bailly, Rigault, Marmagne, Arkoun, Soulay, Caïus, Paysant-Le Roux, Louahlia, Jacquard, Esmaeel, Chardon, Masclaux-Daubresse and Dellagi.
Conflict of interest statement
MA was employed by company TIMAC AGRO International SAS. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Abid M., Tian Z., Ata-Ul-Karim S. T., Cui Y., Liu Y., Zahoor R., et al. (2016). Nitrogen nutrition improves the potential of wheat (Triticum aestivum L.) to alleviate the effects of drought stress during vegetative growth periods. Front. Plant Sci. 7:981. 10.3389/fpls.2016.00981 - DOI - PMC - PubMed
-
- Avila-Ospina L., Marmagne A., Talbotec J., Krupinska K., Masclaux-Daubresse C. (2015). The identification of new cytosolic glutamine synthetase and asparagine synthetase genes in barley (Hordeum vulgare L.), and their expression during leaf senescence. J. Exp. Bot. 66, 2013–2026. 10.1093/jxb/erv003 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

