Comparative Physiology and Transcriptome Analysis of Young Spikes in Response to Late Spring Coldness in Wheat (Triticum aestivum L.)
- PMID: 35185984
- PMCID: PMC8850991
- DOI: 10.3389/fpls.2022.811884
Comparative Physiology and Transcriptome Analysis of Young Spikes in Response to Late Spring Coldness in Wheat (Triticum aestivum L.)
Abstract
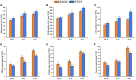
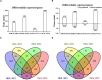
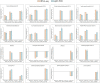
Late spring coldness (LSC) is critical for wheat growth and development in the Huang-Huai valleys of China. However, little is known about the molecular mechanisms for young spikes responding to low temperature (LT) stress during anther connective tissue formation phase (ACFP). To elucidate the molecular mechanisms associated with low temperature, we performed a comparative transcriptome analysis of wheat cultivars Xinmai26 (XM26: cold-sensitive) and Yannong19 (YN19: cold-tolerant) using RNA-seq data. Over 4000 differently expressed genes (DEGs) were identified under low temperature conditions (T1: 4°C) and freezing conditions (T2: -4°C) compared with control (CK: 16°C). The number of DEGs associated with two cultivars at two low temperature treatments (T1: 4°C and T2: -4°C) were 834, 1,353, 231, and 1,882 in four comparison groups (Xinmai26-CK vs. Xinmai26-T1, Xinmai26-CK vs. Xinmai26-T2, Yannong19-CK vs. Yannong19-T1, and Yannong19-CK vs. Yannong19-T2), respectively. Furthermore, to validate the accuracy of RNA-seq, 16 DEGs were analyzed using quantitative real-time RT-PCR. Several transcriptome changes were observed through Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway functional enrichment analysis in plant hormone signal transduction, circadian rhythm-plant, and starch and sucrose metabolism under low temperature. In addition, 126 transcription factors (TFs), including AP2-ERF, bHLH, WRKY, MYB, HSF, and members of the bZIP family, were considered as cold-responsive. It is the first study to investigate DEGs associated with low temperature stress at the transcriptome level in two wheat cultivars with different cold resistance capacities. Most likely, the variations in transcription factors (TFs) regulation, and starch and sucrose metabolism contribute to different cold resistance capacities in the two cultivars. Further, physiological activities of superoxide dismutase (SOD), peroxidase (POD), catalase (CAT) enzymes, malondialdehyde (MDA), soluble sugar (SS), and sucrose contents were evaluated to investigate the negative impacts of low temperature in both cultivars. These findings provide new insight into the molecular mechanisms of plant responses to low temperature and potential candidate genes that required for improving wheat's capacity to withstand low temperature stress.
Keywords: Triticum aestivum L.; anther connective tissue formation phase (ACFP); differentially expressed genes (DEGs); late spring coldness (LSC); physiology and transcriptome.
Copyright © 2022 Jiang, Hassan, Muhammad, Arshad, Chen, Xu, Xu, Ni, Liu, Yang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

