Human/SARS-CoV-2 genome-scale metabolic modeling to discover potential antiviral targets for COVID-19
- PMID: 35186172
- PMCID: PMC8843340
- DOI: 10.1016/j.jtice.2022.104273
Human/SARS-CoV-2 genome-scale metabolic modeling to discover potential antiviral targets for COVID-19
Abstract
Background: Coronavirus disease 2019 (COVID-19) has caused a substantial increase in mortality and economic and social disruption. The absence of US Food and Drug Administration-approved drugs for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) highlights the need for new therapeutic drugs to combat COVID-19.
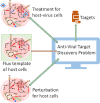
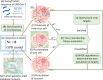
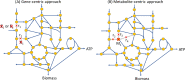
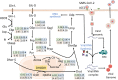
Methods: The present study proposed a fuzzy hierarchical optimization framework for identifying potential antiviral targets for COVID-19. The objectives in the decision-making problem were not only to evaluate the elimination of the virus growth, but also to minimize side effects causing treatment. The identified candidate targets could promote processes of drug discovery and development.
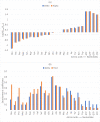
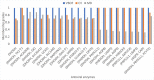
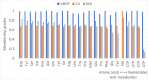
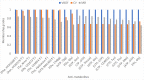
Significant findings: Our gene-centric method revealed that dihydroorotate dehydrogenase (DHODH) inhibition could reduce viral biomass growth and metabolic deviation by 99.4% and 65.6%, respectively, and increase cell viability by 70.4%. We also identified two-target combinations that could completely block viral biomass growth and more effectively prevent metabolic deviation. We also discovered that the inhibition of two antiviral metabolites, cytidine triphosphate (CTP) and uridine-5'-triphosphate (UTP), exhibits effects similar to those of molnupiravir, which is undergoing phase III clinical trials. Our predictions also indicate that CTP and UTP inhibition blocks viral RNA replication through a similar mechanism to that of molnupiravir.
Keywords: Bioprocess systems engineering; Computer-aided drug discovery; Constraint-based modeling; Evolutionary optimization; Flux balance analysis; Fuzzy optimization.
© 2022 Taiwan Institute of Chemical Engineers. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
Cell-specific genome-scale metabolic modeling of SARS-CoV-2-infected lung to identify antiviral enzymes.FEBS Open Bio. 2023 Dec;13(12):2172-2186. doi: 10.1002/2211-5463.13710. Epub 2023 Sep 30. FEBS Open Bio. 2023. PMID: 37734920 Free PMC article.
-
Fuzzy optimization for identifying antiviral targets for treating SARS-CoV-2 infection in the heart.BMC Bioinformatics. 2023 Sep 27;24(1):364. doi: 10.1186/s12859-023-05487-7. BMC Bioinformatics. 2023. PMID: 37759157 Free PMC article.
-
Molnupiravir and Its Antiviral Activity Against COVID-19.Front Immunol. 2022 Apr 4;13:855496. doi: 10.3389/fimmu.2022.855496. eCollection 2022. Front Immunol. 2022. PMID: 35444647 Free PMC article. Review.
-
Current Strategies of Antiviral Drug Discovery for COVID-19.Front Mol Biosci. 2021 May 13;8:671263. doi: 10.3389/fmolb.2021.671263. eCollection 2021. Front Mol Biosci. 2021. PMID: 34055887 Free PMC article. Review.
-
Inhibitors of dihydroorotate dehydrogenase cooperate with molnupiravir and N4-hydroxycytidine to suppress SARS-CoV-2 replication.iScience. 2022 May 20;25(5):104293. doi: 10.1016/j.isci.2022.104293. Epub 2022 Apr 25. iScience. 2022. PMID: 35492218 Free PMC article.
Cited by
-
Discovery of a New Class of Thiazolidin-4-one-Based Inhibitors of Human Dihydroorotate Dehydrogenase: Biological Activity Evaluation, Molecular Docking, and Molecular Dynamics.ACS Omega. 2025 Mar 21;10(12):12393-12402. doi: 10.1021/acsomega.4c11459. eCollection 2025 Apr 1. ACS Omega. 2025. PMID: 40191330 Free PMC article.
-
Cell-specific genome-scale metabolic modeling of SARS-CoV-2-infected lung to identify antiviral enzymes.FEBS Open Bio. 2023 Dec;13(12):2172-2186. doi: 10.1002/2211-5463.13710. Epub 2023 Sep 30. FEBS Open Bio. 2023. PMID: 37734920 Free PMC article.
-
Context-Specific Genome-Scale Metabolic Modelling and Its Application to the Analysis of COVID-19 Metabolic Signatures.Metabolites. 2023 Jan 13;13(1):126. doi: 10.3390/metabo13010126. Metabolites. 2023. PMID: 36677051 Free PMC article. Review.
-
Fuzzy optimization for identifying antiviral targets for treating SARS-CoV-2 infection in the heart.BMC Bioinformatics. 2023 Sep 27;24(1):364. doi: 10.1186/s12859-023-05487-7. BMC Bioinformatics. 2023. PMID: 37759157 Free PMC article.
References
-
- Chakraborty C., Sharma A.R., Sharma G., Bhattacharya M., Lee S.S. SARS-CoV-2 causing pneumonia-associated respiratory disorder (COVID-19): diagnostic and proposed therapeutic options. Eur Rev Med Pharmacol Sci. 2020;24(7):4016–4026. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
