The history of foot-and-mouth disease virus serotype C: the first known extinct serotype?
- PMID: 35186323
- PMCID: PMC8102019
- DOI: 10.1093/ve/veab009
The history of foot-and-mouth disease virus serotype C: the first known extinct serotype?
Abstract
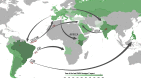
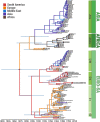
Foot-and-mouth disease (FMD) is a highly contagious animal disease caused by an RNA virus subdivided into seven serotypes that are unevenly distributed in Asia, Africa, and South America. Despite the challenges of controlling FMD, since 1996 there have been only two outbreaks attributed to serotype C, in Brazil and in Kenya, in 2004. This article describes the historical distribution and origins of serotype C and its disappearance. The serotype was first described in Europe in the 1920s, where it mainly affected pigs and cattle but as a less common cause of outbreaks than serotypes O and A. No serotype C outbreaks have been reported in Europe since vaccination stopped in 1990. FMD virus is presumed to have been introduced into South America from Europe in the nineteenth century, although whether serotype C evolved there or in Europe is not known. As in Europe, this serotype was less widely distributed and caused fewer outbreaks than serotypes O and A. Since 1994, serotype C had not been reported from South America until four small outbreaks were detected in the Amazon region in 2004. Elsewhere, serotype C was introduced to Asia, in the 1950s to the 1970s, persisting and evolving for several decades in the Indian subcontinent and for eighteen years in the Philippines. Serotype C virus also circulated in East Africa between 1957 and 2004. Many serotype C viruses from European and Kenyan outbreaks were closely related to vaccine strains, including the most recently recovered Kenyan isolate from 2004. International surveillance has not confirmed any serotype C cases, worldwide, for over 15 years, despite more than 2,000 clinical submissions per year to reference laboratories. Serology provides limited evidence for absence of this serotype, as unequivocal interpretation is hampered by incomplete intra-serotype specificity of immunoassays and the continued use of this serotype in vaccines. It is recommended to continue strengthening surveillance in regions of FMD endemicity, to stop vaccination against serotype C and to reduce working with the virus in laboratories, since inadvertent escape of virus during such activities is now the biggest risk for its reappearance in the field.
Keywords: extinction; foot-and-mouth; phylogeny; serotype C.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures

References
Publication types
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

