T-Cell Subpopulations Exhibit Distinct Recruitment Potential, Immunoregulatory Profile and Functional Characteristics in Chagas versus Idiopathic Dilated Cardiomyopathies
- PMID: 35187122
- PMCID: PMC8847602
- DOI: 10.3389/fcvm.2022.787423
T-Cell Subpopulations Exhibit Distinct Recruitment Potential, Immunoregulatory Profile and Functional Characteristics in Chagas versus Idiopathic Dilated Cardiomyopathies
Abstract
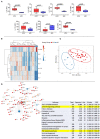
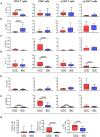
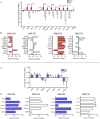
Chronic Chagas cardiomyopathy (CCC) is one of the deadliest cardiomyopathies known and the most severe manifestation of Chagas disease, which is caused by infection with the parasite Trypanosoma cruzi. Idiopathic dilated cardiomyopathies (IDC) are a diverse group of inflammatory heart diseases that affect the myocardium and are clinically similar to CCC, often causing heart failure and death. While T-cells are critical for mediating cardiac pathology in CCC and IDC, the mechanisms underlying T-cell function in these cardiomyopathies are not well-defined. In this study, we sought to investigate the phenotypic and functional characteristics of T-cell subpopulations in CCC and IDC, aiming to clarify whether the inflammatory response is similar or distinct in these cardiomyopathies. We evaluated the expression of systemic cytokines, determined the sources of the different cytokines, the expression of their receptors, of cytotoxic molecules, and of molecules associated with recruitment to the heart by circulating CD4+, CD8+, and CD4-CD8- T-cells from CCC and IDC patients, using multiparameter flow cytometry combined with conventional and unsupervised machine-learning strategies. We also used an in silico approach to identify the expression of genes that code for key molecules related to T-cell function in hearts of patient with CCC and IDC. Our data demonstrated that CCC patients displayed a more robust systemic inflammatory cytokine production as compared to IDC. While CD8+ T-cells were highly activated in CCC as compared to IDC, CD4+ T-cells were more activated in IDC. In addition to differential expression of functional molecules, these cells also displayed distinct expression of molecules associated with recruitment to the heart. In silico analysis of gene transcripts in the cardiac tissue demonstrated a significant correlation between CD8 and inflammatory, cytotoxic and cardiotropic molecules in CCC transcripts, while no correlation with CD4 was observed. A positive correlation was observed between CD4 and perforin transcripts in hearts from IDC but not CCC, as compared to normal tissue. These data show a clearly distinct systemic and local cellular response in CCC and IDC, despite their similar cardiac impairment, which may contribute to identifying specific immunotherapeutic targets in these diseases.
Keywords: Chagas cardiomyopathy; T-cells; chemokines; cytokines; idiopathic cardiomyopathy; inflammation.
Copyright © 2022 Neves, Koh, Souza-Silva, Passos, Silva, Velikkakam, Villani, Coelho, Brodskyn, Teixeira, Gollob, Nunes and Dutra.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer JS declared a shared affiliation with one of the authors AT, to the handling editor at time of review.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials

