Individual variation in susceptibility or exposure to SARS-CoV-2 lowers the herd immunity threshold
- PMID: 35189135
- PMCID: PMC8855661
- DOI: 10.1016/j.jtbi.2022.111063
Individual variation in susceptibility or exposure to SARS-CoV-2 lowers the herd immunity threshold
Abstract
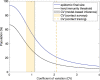
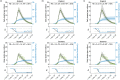
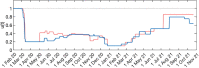
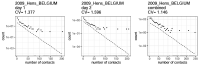
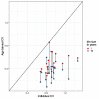
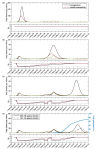
Individual variation in susceptibility and exposure is subject to selection by natural infection, accelerating the acquisition of immunity, and reducing herd immunity thresholds and epidemic final sizes. This is a manifestation of a wider population phenomenon known as "frailty variation". Despite theoretical understanding, public health policies continue to be guided by mathematical models that leave out considerable variation and as a result inflate projected disease burdens and overestimate the impact of interventions. Here we focus on trajectories of the coronavirus disease (COVID-19) pandemic in England and Scotland until November 2021. We fit models to series of daily deaths and infer relevant epidemiological parameters, including coefficients of variation and effects of non-pharmaceutical interventions which we find in agreement with independent empirical estimates based on contact surveys. Our estimates are robust to whether the analysed data series encompass one or two pandemic waves and enable projections compatible with subsequent dynamics. We conclude that vaccination programmes may have contributed modestly to the acquisition of herd immunity in populations with high levels of pre-existing naturally acquired immunity, while being crucial to protect vulnerable individuals from severe outcomes as the virus becomes endemic.
Keywords: COVID-19; Epidemic model; Frailty variation; Herd immunity threshold; Individual variation; Selection within cohorts.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
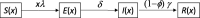




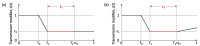





Update of
-
Individual variation in susceptibility or exposure to SARS-CoV-2 lowers the herd immunity threshold.medRxiv [Preprint]. 2022 Feb 14:2020.04.27.20081893. doi: 10.1101/2020.04.27.20081893. medRxiv. 2022. Update in: J Theor Biol. 2022 May 7;540:111063. doi: 10.1016/j.jtbi.2022.111063. PMID: 32511451 Free PMC article. Updated. Preprint.
References
-
- Aalen O.O. Heterogeneity in survival analysis. Stat. Med. 1988;7:1121–1137. - PubMed
-
- Adam D.C., Wu P., Wong J.Y., Lau E.H.Y., Tsang T.K., Cauchemez S., et al. Clustering and superspreading potential of SARS-CoV-2 infections in Hong Kong. Nat. Med. 2020;26:1714–1719. - PubMed
-
- Aguas R., Corder R.M., King J.G., Gonçalves G., Ferreira M.U., Gomes M.G.M. Herd immunity thresholds for SARS-CoV-2 estimated from unfolding epidemics. Preprint medRxiv. 2020 doi: 10.1101/2020.07.23.20160762. - DOI
-
- Althaus C.L., Baggio S., Reichmuth M.L., Hodcroft E.B., Riou J., Neher R.A., et al. A tale of two variants: Spread of SARS-CoV-2 variants Alpha in Geneva, Switzerland, and Beta in South Africa. Preprint medRxiv. 2021 doi: 10.1101/2021.06.10.21258468. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

