Molecular dissection of RbpA-mediated regulation of fidaxomicin sensitivity in mycobacteria
- PMID: 35189142
- PMCID: PMC8956947
- DOI: 10.1016/j.jbc.2022.101752
Molecular dissection of RbpA-mediated regulation of fidaxomicin sensitivity in mycobacteria
Abstract
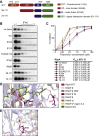
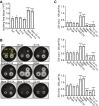
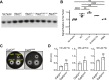
RNA polymerase (RNAP) binding protein A (RbpA) is essential for mycobacterial viability and regulates transcription initiation by increasing the stability of the RNAP-promoter open complex (RPo). RbpA consists of four domains: an N-terminal tail (NTT), a core domain (CD), a basic linker, and a sigma interaction domain. We have previously shown that truncation of the RbpA NTT and CD increases RPo stabilization by RbpA, implying that these domains inhibit this activity of RbpA. Previously published structural studies showed that the NTT and CD are positioned near multiple RNAP-σA holoenzyme functional domains and predict that the RbpA NTT contributes specific amino acids to the binding site of the antibiotic fidaxomicin (Fdx), which inhibits the formation of the RPo complex. Furthermore, deletion of the NTT results in decreased Mycobacterium smegmatis sensitivity to Fdx, but whether this is caused by a loss in Fdx binding is unknown. We generated a panel of rbpA mutants and found that the RbpA NTT residues predicted to directly interact with Fdx are partially responsible for RbpA-dependent Fdx activity in vitro, while multiple additional RbpA domains contribute to Fdx activity in vivo. Specifically, our results suggest that the RPo-stabilizing activity of RbpA decreases Fdx activity in vivo. In support of the association between RPo stability and Fdx activity, we find that another factor that promotes RPo stability in bacteria, CarD, also impacts to Fdx sensitivity. Our findings highlight how RbpA and other factors may influence RNAP dynamics to affect Fdx sensitivity.
Keywords: RNA polymerase; RbpA; antibiotic action; bacterial genetics; bacterial transcription; fidaxomicin; mycobacteria.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

