Nucleic acid aptamer controls mycoplasma infection for inhibiting the malignancy of esophageal squamous cell carcinoma
- PMID: 35189346
- PMCID: PMC9171152
- DOI: 10.1016/j.ymthe.2022.02.018
Nucleic acid aptamer controls mycoplasma infection for inhibiting the malignancy of esophageal squamous cell carcinoma
Abstract
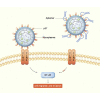
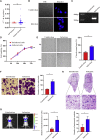
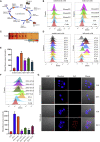
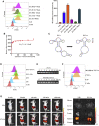
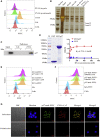
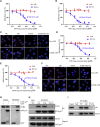
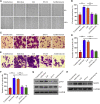
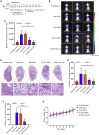
Esophageal cancer is one of the most frequent malignant tumors of the digestive tract, among which esophageal squamous cell carcinoma (ESCC) is the main pathological type worldwide. Previous studies have shown microbial infections in the upper digestive tract to be a potential risk factor in ESCC etiology. In this study, we identified that Mycoplasma hyorhinis infection promoted the malignancy of ESCC. In response, we generated a single-stranded DNA aptamer, ZY3A, against M. hyorhinis using the cell-SELEX strategy. The underlying recognition mechanism of ZY3A on M. hyorhinis involves its binding to M. hyorhinis-specific p37 protein. This tool allowed us to provide the first proof-of-concept evidence using a nucleic acid aptamer to control mycoplasma infection. More specifically, we found that ZY3A could neutralize M. hyorhinis infection on ESCC cells by blocking the interaction between p37 protein and its receptor TLR4 on the ESCC cell membrane. As a result, ZY3A inhibited the migration and invasion of M. hyorhinis-infected ESCC cells in vitro and metastasis in vivo. Taken together, these findings indicate that aptamer ZY3A is a potential candidate for development into a novel molecular tool for treatment of M. hyorhinis infection and a safe first-in-class M. hyorhinis-targeting antitumor agent.
Keywords: M. hyorhinis; esophageal squamous cell carcinoma; nucleic acid aptamer; p37 protein; tumor metastasis.
Copyright © 2022 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare that they have no competing interests.
Figures
Similar articles
-
Mycoplasma hyorhinis infection promotes NF-κB-dependent migration of gastric cancer cells.Cancer Res. 2014 Oct 15;74(20):5782-94. doi: 10.1158/0008-5472.CAN-14-0650. Epub 2014 Aug 18. Cancer Res. 2014. PMID: 25136068
-
Mycoplasma hyorhinis infection in gastric carcinoma and its effects on the malignant phenotypes of gastric cancer cells.BMC Gastroenterol. 2010 Nov 10;10:132. doi: 10.1186/1471-230X-10-132. BMC Gastroenterol. 2010. PMID: 21062494 Free PMC article.
-
Intratumor Mycoplasma promotes the initiation and progression of hepatocellular carcinoma.Cell Rep. 2023 Dec 26;42(12):113563. doi: 10.1016/j.celrep.2023.113563. Epub 2023 Dec 12. Cell Rep. 2023. PMID: 38088929
-
Mycoplasma hyorhinis infection promotes gastric cancer cell motility via β-catenin signaling.Cancer Med. 2019 Sep;8(11):5301-5312. doi: 10.1002/cam4.2357. Epub 2019 Jul 18. Cancer Med. 2019. PMID: 31321908 Free PMC article.
-
Mycoplasma infection promotes tumor progression via interaction of the mycoplasmal protein p37 and epithelial cell adhesion molecule in hepatocellular carcinoma.Cancer Lett. 2019 Jul 10;454:44-52. doi: 10.1016/j.canlet.2019.04.007. Epub 2019 Apr 10. Cancer Lett. 2019. PMID: 30980864
Cited by
-
A Milestone in the Shift from "Passive Killing" to "Active Immunomodulation" in Cancer Treatment-Progress in Melanoma Vaccine Research.Curr Treat Options Oncol. 2025 Jul 17. doi: 10.1007/s11864-025-01340-6. Online ahead of print. Curr Treat Options Oncol. 2025. PMID: 40676479 Review.
References
-
- Ajani J.A., D'Amico T.A., Bentrem D.J., Chao J., Corvera C., Das P., Denlinger C.S., Enzinger P.C., Fanta P., Farjah F., et al. Esophageal and esophagogastric junction cancers, version 2.2019. Natl. Comp. Canc. Netw. 2019;17:855–883. - PubMed
-
- Huang F.L., Yu S.J. Esophageal cancer: risk factors, genetic association, and treatment. Asian J. Surg. 2018;41:210–215. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

