Cancer/Testis Antigen 55 is required for cancer cell proliferation and mitochondrial DNA maintenance
- PMID: 35189384
- PMCID: PMC9057655
- DOI: 10.1016/j.mito.2022.02.005
Cancer/Testis Antigen 55 is required for cancer cell proliferation and mitochondrial DNA maintenance
Abstract
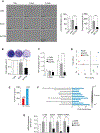
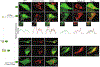
Cancer/Testis Antigens (CTAs) represent a group of proteins whose expression under physiological conditions is restricted to testis but activated in many human cancers. Also, it was observed that co-expression of multiple CTAs worsens the patient prognosis. Five CTAs were reported acting in mitochondria and we recently reported 147 transcripts encoded by 67 CTAs encoding for proteins potentially targeted to mitochondria. Among them, we identified the two isoforms encoded by CT55 for whom the function is poorly understood. First, we found that patients with tumors expressing wild-type CT55 are associated with poor survival. Moreover, CT55 silencing decreases dramatically cell proliferation. Second, to investigate the role of CT55 on mitochondria, we first show that CT55 is localized to both mitochondria and endoplasmic reticulum (ER) due to the presence of an ambiguous N-terminal targeting signal. Then, we show that CT55 silencing decreases mtDNA copy number and delays mtDNA recovery after an acute depletion. Moreover, demethylation of CT55 promotor increases its expression, which in turn increases mtDNA copy number. Finally, we measured the mtDNA copy number in NCI-60 cell lines and screened for genes whose expression is strongly correlated to mtDNA amount. We identified CT55 as the second highest correlated hit. Also, we show that compared to siRNA scrambled control (siCtrl) treatment, CT55 specific siRNA (siCT55) treatment down-regulates aerobic respiration, indicating that CT55 sustains mitochondrial respiration. Altogether, these data show for first time that CT55 acts on mtDNA copy number, modulates mitochondrial activity to sustain cancer cell proliferation.
Keywords: CT55; Cell proliferation; Mitochondrial DNA; NCI-60.
Copyright © 2022 Elsevier B.V. and Mitochondria Research Society. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
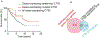
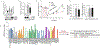
References
-
- van der Bruggen P et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 254, 1643–1647 (1991). - PubMed
-
- Hüttemann M, Jaradat S & Grossman LI Cytochrome c oxidase of mammals contains a testes-specific isoform of subunit VIb-the counterpart to testes-specific cytochrome c?: COX VIb TESTES. Mol. Reprod. Dev. 66, 8–16 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

