NBS1-CtIP-mediated DNA end resection suppresses cGAS binding to micronuclei
- PMID: 35189637
- PMCID: PMC8934670
- DOI: 10.1093/nar/gkac079
NBS1-CtIP-mediated DNA end resection suppresses cGAS binding to micronuclei
Abstract
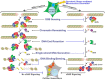
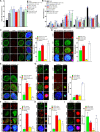
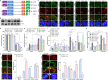
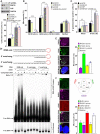
Cyclic guanosine monophosphate-adenosine monophosphate synthase (cGAS) is activated in cells with defective DNA damage repair and signaling (DDR) factors, but a direct role for DDR factors in regulating cGAS activation in response to micronuclear DNA is still poorly understood. Here, we provide novel evidence that Nijmegen breakage syndrome 1 (NBS1) protein, a well-studied DNA double-strand break (DSB) sensor-in coordination with Ataxia Telangiectasia Mutated (ATM), a protein kinase, and Carboxy-terminal binding protein 1 interacting protein (CtIP), a DNA end resection factor-functions as an upstream regulator that prevents cGAS from binding micronuclear DNA. When NBS1 binds to micronuclear DNA via its fork-head-associated domain, it recruits CtIP and ATM via its N- and C-terminal domains, respectively. Subsequently, ATM stabilizes NBS1's interaction with micronuclear DNA, and CtIP converts DSB ends into single-strand DNA ends; these two key events prevent cGAS from binding micronuclear DNA. Additionally, by using a cGAS tripartite system, we show that cells lacking NBS1 not only recruit cGAS to a major fraction of micronuclear DNA but also activate cGAS in response to these micronuclear DNA. Collectively, our results underscore how NBS1 and its binding partners prevent cGAS from binding micronuclear DNA, in addition to their classical functions in DDR signaling.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

