Inferring person-to-person networks of Plasmodium falciparum transmission: are analyses of routine surveillance data up to the task?
- PMID: 35189905
- PMCID: PMC8860266
- DOI: 10.1186/s12936-022-04072-2
Inferring person-to-person networks of Plasmodium falciparum transmission: are analyses of routine surveillance data up to the task?
Abstract
Background: Inference of person-to-person transmission networks using surveillance data is increasingly used to estimate spatiotemporal patterns of pathogen transmission. Several data types can be used to inform transmission network inferences, yet the sensitivity of those inferences to different data types is not routinely evaluated.
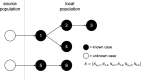
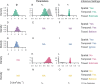
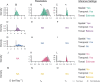
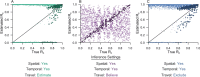
Methods: The influence of different combinations of spatial, temporal, and travel-history data on transmission network inferences for Plasmodium falciparum malaria were evaluated.
Results: The information content of these data types may be limited for inferring person-to-person transmission networks and may lead to an overestimate of transmission. Only when outbreaks were temporally focal or travel histories were accurate was the algorithm able to accurately estimate the reproduction number under control, Rc. Applying this approach to data from Eswatini indicated that inferences of Rc and spatiotemporal patterns therein depend upon the choice of data types and assumptions about travel-history data.
Conclusions: These results suggest that transmission network inferences made with routine malaria surveillance data should be interpreted with caution.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures