Genomic Correlates of Outcome in Tumor-Infiltrating Lymphocyte Therapy for Metastatic Melanoma
- PMID: 35190823
- PMCID: PMC9064946
- DOI: 10.1158/1078-0432.CCR-21-1060
Genomic Correlates of Outcome in Tumor-Infiltrating Lymphocyte Therapy for Metastatic Melanoma
Abstract
Purpose: Adoptive cell therapy (ACT) of tumor-infiltrating lymphocytes (TIL) historically yields a 40%-50% response rate in metastatic melanoma. However, the determinants of outcome are largely unknown.
Experimental design: We investigated tumor-based genomic correlates of overall survival (OS), progression-free survival (PFS), and response to therapy by interrogating tumor samples initially collected to generate TIL infusion products.
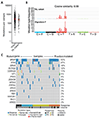
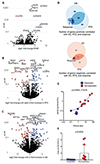
Results: Whole-exome sequencing (WES) data from 64 samples indicated a positive correlation between neoantigen load and OS, but not PFS or response to therapy. RNA sequencing analysis of 34 samples showed that expression of PDE1C, RTKN2, and NGFR was enriched in responders who had improved PFS and OS. In contrast, the expression of ELFN1 was enriched in patients with unfavorable response, poor PFS and OS, whereas enhanced methylation of ELFN1 was observed in patients with favorable outcomes. Expression of ELFN1, NGFR, and PDE1C was mainly found in cancer-associated fibroblasts and endothelial cells in tumor tissues across different cancer types in publicly available single-cell RNA sequencing datasets, suggesting a role for elements of the tumor microenvironment in defining the outcome of TIL therapy.
Conclusions: Our findings suggest that transcriptional features of melanomas correlate with outcomes after TIL therapy and may provide candidates to guide patient selection.
©2022 American Association for Cancer Research.
Figures


References
-
- Goff SL, Dudley ME, Citrin DE, Somerville RP, Wunderlich JR, Danforth DN, et al. Randomized, Prospective Evaluation Comparing Intensity of Lymphodepletion Before Adoptive Transfer of Tumor-Infiltrating Lymphocytes for Patients With Metastatic Melanoma. J Clin Oncol 2016;34(20):2389–97 doi 10.1200/JCO.2016.66.7220. - DOI - PMC - PubMed
-
- Andersen R, Donia M, Ellebaek E, Borch TH, Kongsted P, Iversen TZ, et al. Long-Lasting Complete Responses in Patients with Metastatic Melanoma after Adoptive Cell Therapy with Tumor-Infiltrating Lymphocytes and an Attenuated IL2 Regimen. Clin Cancer Res 2016;22(15):3734–45 doi 10.1158/1078-0432.CCR-15-1879. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

