Renal tubular peroxisomes are dispensable for normal kidney function
- PMID: 35191396
- PMCID: PMC8876468
- DOI: 10.1172/jci.insight.155836
Renal tubular peroxisomes are dispensable for normal kidney function
Abstract
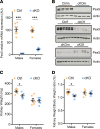
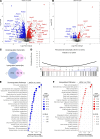
Peroxisomes are specialized cellular organelles involved in a variety of metabolic processes. In humans, mutations leading to complete loss of peroxisomes cause multiorgan failure (Zellweger's spectrum disorders, ZSD), including renal impairment. However, the (patho)physiological role of peroxisomes in the kidney remains unknown. We addressed the role of peroxisomes in renal function in mice with conditional ablation of peroxisomal biogenesis in the renal tubule (cKO mice). Functional analyses did not reveal any overt kidney phenotype in cKO mice. However, infant male cKO mice had lower body and kidney weights, and adult male cKO mice exhibited substantial reductions in kidney weight and kidney weight/body weight ratio. Stereological analysis showed an increase in mitochondria density in proximal tubule cells of cKO mice. Integrated transcriptome and metabolome analyses revealed profound reprogramming of a number of metabolic pathways, including metabolism of glutathione and biosynthesis/biotransformation of several major classes of lipids. Although this analysis suggested compensated oxidative stress, challenge with high-fat feeding did not induce significant renal impairments in cKO mice. We demonstrate that renal tubular peroxisomes are dispensable for normal renal function. Our data also suggest that renal impairments in patients with ZSD are of extrarenal origin.
Keywords: Genetic diseases; Nephrology.
Figures





References
-
- Rhodin JAG. Correlation of ultrastructural oganisation and function in normal and experimentally changed proximal tubule cells of the mouse kidney: an electron microscopic study. PhD Thesis. Karolinska Institutet; 1954.
-
- Reddy JK, et al. Microbody (peroxisome) proliferation in mouse kidney induced by methyl clofenapate. Virchows Arch B Cell Pathol. 1975;17(4):295–306. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

