Development of an immunofluorescence assay for detection of SARS-CoV-2
- PMID: 35192015
- PMCID: PMC8862410
- DOI: 10.1007/s00705-022-05392-z
Development of an immunofluorescence assay for detection of SARS-CoV-2
Abstract
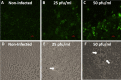
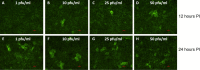
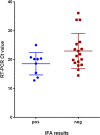
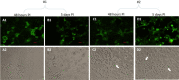
SARS-CoV-2, the etiologic agent of the COVID-19 pandemic, emerged as the cause of a global crisis in 2019. Currently, the main method for identification of SARS-CoV-2 is a reverse transcription (RT)-PCR assay designed to detect viral RNA in oropharyngeal (OP) or nasopharyngeal (NP) samples. While the PCR assay is considered highly specific and sensitive, this method cannot determine the infectivity of the sample, which may assist in evaluation of virus transmissibility from patients and breaking transmission chains. Thus, cell-culture-based approaches such as cytopathic effect (CPE) assays are routinely employed for the identification of infectious viruses in NP/OP samples. Despite their high sensitivity, CPE assays take several days and require additional diagnostic tests in order to verify the identity of the pathogen. We have therefore developed a rapid immunofluorescence assay (IFA) for the specific detection of SARS-CoV-2 in NP/OP samples following cell culture infection. Initially, IFA was carried out on Vero E6 cultures infected with SARS-CoV-2 at defined concentrations, and infection was monitored at different time points. This test was able to yield positive signals in cultures infected with 10 pfu/ml at 12 hours postinfection (PI). Increasing the incubation time to 24 hours reduced the detectable infective dose to 1 pfu/ml. These IFA signals occur before the development of CPE. When compared to the CPE test, IFA has the advantages of specificity, rapid detection, and sensitivity, as demonstrated in this work.
© 2022. The Author(s), under exclusive licence to Springer-Verlag GmbH Austria, part of Springer Nature.
Conflict of interest statement
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

