Histone demethylase IBM1-mediated meiocyte gene expression ensures meiotic chromosome synapsis and recombination
- PMID: 35192603
- PMCID: PMC8896719
- DOI: 10.1371/journal.pgen.1010041
Histone demethylase IBM1-mediated meiocyte gene expression ensures meiotic chromosome synapsis and recombination
Abstract
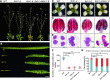
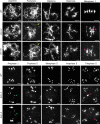
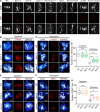
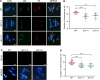
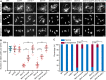
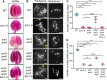
Histone methylation and demethylation play important roles in plant growth and development, but the involvement of histone demethylation during meiosis is poorly understood. Here we show that disruption of Arabidopsis thaliana INCREASE IN BONSAI METHYLATION 1 (IBM1) causes incomplete synapsis, chromosome entanglement and reduction of recombination during meiosis, leading to sterility. Interestingly, these ibm1 meiotic defects are rescued by mutations in either SUVH4/KYP or CMT3. Using transcriptomic analyses we show that mutation of IBM1 down-regulates thousands of genes expressed in meiocytes, and that expression of about 38% of these genes are restored to wild type levels in ibm1 cmt3 double mutants. Changes in the expression of 437 of these, including the ARABIDOPSIS MEI2-LIKE AML3-5 genes, are correlated with a significant reduction of gene body CHG methylation. Consistently, the aml3 aml4 aml5 triple have defects in synapsis and chromosome entanglement similar to ibm1. Genetic analysis shows that aml3 aml4 aml5 ibm1 quadruple mutants resembles the ibm1 single mutant. Strikingly, over expression of AML5 in ibm1 can partially rescue the ibm1 meiotic defects. Taken together, our results demonstrate that histone demethylase IBM1 is required for meiosis likely via coordinated regulation of meiocyte gene expression during meiosis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Dynamic evolution of the heterochromatin sensing histone demethylase IBM1.PLoS Genet. 2024 Jul 11;20(7):e1011358. doi: 10.1371/journal.pgen.1011358. eCollection 2024 Jul. PLoS Genet. 2024. PMID: 38991029 Free PMC article.
-
Autocatalytic differentiation of epigenetic modifications within the Arabidopsis genome.EMBO J. 2010 Oct 20;29(20):3496-506. doi: 10.1038/emboj.2010.227. Epub 2010 Sep 10. EMBO J. 2010. PMID: 20834229 Free PMC article.
-
Demethylation of ERECTA receptor genes by IBM1 histone demethylase affects stomatal development.Development. 2016 Dec 1;143(23):4452-4461. doi: 10.1242/dev.129932. Epub 2016 Oct 3. Development. 2016. PMID: 27697902 Free PMC article.
-
Meiotic chromosome synapsis and recombination in Arabidopsis thaliana: new ways of integrating cytological and molecular approaches.Chromosome Res. 2014 Jun;22(2):179-90. doi: 10.1007/s10577-014-9426-8. Chromosome Res. 2014. PMID: 24941912 Review.
-
Meiotic chromosome synapsis and recombination in Arabidopsis thaliana; an integration of cytological and molecular approaches.Chromosome Res. 2003;11(3):205-15. doi: 10.1023/a:1022831724990. Chromosome Res. 2003. PMID: 12769288 Review.
Cited by
-
Dynamic evolution of the heterochromatin sensing histone demethylase IBM1.PLoS Genet. 2024 Jul 11;20(7):e1011358. doi: 10.1371/journal.pgen.1011358. eCollection 2024 Jul. PLoS Genet. 2024. PMID: 38991029 Free PMC article.
-
DNA Methylation and Alternative Splicing Safeguard Genome and Transcriptome After a Retrotransposition Burst in Arabidopsis thaliana.Int J Mol Sci. 2025 May 17;26(10):4816. doi: 10.3390/ijms26104816. Int J Mol Sci. 2025. PMID: 40429956 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

