The EurOPDX Data Portal: an open platform for patient-derived cancer xenograft data sharing and visualization
- PMID: 35193494
- PMCID: PMC8862363
- DOI: 10.1186/s12864-022-08367-1
The EurOPDX Data Portal: an open platform for patient-derived cancer xenograft data sharing and visualization
Abstract
Background: Patient-derived xenografts (PDX) mice models play an important role in preclinical trials and personalized medicine. Sharing data on the models is highly valuable for numerous reasons - ethical, economical, research cross validation etc. The EurOPDX Consortium was established 8 years ago to share such information and avoid duplicating efforts in developing new PDX mice models and unify approaches to support preclinical research. EurOPDX Data Portal is the unified data sharing platform adopted by the Consortium.
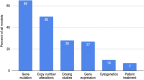
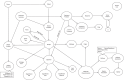
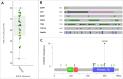
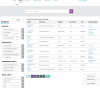
Main body: In this paper we describe the main features of the EurOPDX Data Portal ( https://dataportal.europdx.eu/ ), its architecture and possible utilization by researchers who look for PDX mice models for their research. The Portal offers a catalogue of European models accessible on a cooperative basis. The models are searchable by metadata, and a detailed view provides molecular profiles (gene expression, mutation, copy number alteration) and treatment studies. The Portal displays the data in multiple tools (PDX Finder, cBioPortal, and GenomeCruzer in future), which are populated from a common database displaying strictly mutually consistent views.
(short) conclusion: EurOPDX Data Portal is an entry point to the EurOPDX Research Infrastructure offering PDX mice models for collaborative research, (meta)data describing their features and deep molecular data analysis according to users' interests.
Keywords: Data harmonization; Database; Molecular data analysis; PDX; Research infrastructure.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Byrne AT, Alférez DG, Amant F, Annibali D, Arribas J, Biankin AV, Bruna A, Budinská E, Caldas C, Chang DK, Clarke RB, Clevers H, Coukos G, Dangles-Marie V, Eckhardt SG, Gonzalez-Suarez E, Hermans E, Hidalgo M, Jarzabek MA, de Jong S, Jonkers J, Kemper K, Lanfrancone L, Mælandsmo GM, Marangoni E, Marine JC, Medico E, Norum JH, Palmer HG, Peeper DS, Pelicci PG, Piris-Gimenez A, Roman-Roman S, Rueda OM, Seoane J, Serra V, Soucek L, Vanhecke D, Villanueva A, Vinolo E, Bertotti A, Trusolino L. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat Rev Cancer. 2017;17(4):254–268. doi: 10.1038/nrc.2016.140. - DOI - PubMed
-
- Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F, Zagonel V, Leone F, Depetris I, Martinelli E, Troiani T, Ciardiello F, Racca P, Bertotti A, Siravegna G, Torri V, Amatu A, Ghezzi S, Marrapese G, Palmeri L, Valtorta E, Cassingena A, Lauricella C, Vanzulli A, Regge D, Veronese S, Comoglio PM, Bardelli A, Marsoni S, Siena S. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17(6):738–746. doi: 10.1016/S1470-2045(16)00150-9. - DOI - PubMed
-
- Meehan TF, Conte N, Goldstein T, Inghirami G, Murakami MA, Brabetz S, Gu Z, Wiser JA, Dunn P, Begley DA, Krupke DM, Bertotti A, Bruna A, Brush MH, Byrne AT, Caldas C, Christie AL, Clark DA, Dowst H, Dry JR, Doroshow JH, Duchamp O, Evrard YA, Ferretti S, Frese KK, Goodwin NC, Greenawalt D, Haendel MA, Hermans E, Houghton PJ, Jonkers J, Kemper K, Khor TO, Lewis MT, Lloyd KCK, Mason J, Medico E, Neuhauser SB, Olson JM, Peeper DS, Rueda OM, Seong JK, Trusolino L, Vinolo E, Wechsler-Reya RJ, Weinstock DM, Welm A, Weroha SJ, Amant F, Pfister SM, Kool M, Parkinson H, Butte AJ, Bult CJ. PDX-MI: minimal information for patient-derived tumor xenograft models. Cancer Res. 2017;77(21):e62–e66. doi: 10.1158/0008-5472.CAN-17-0582. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

