Cooperative multivalent receptor binding promotes exposure of the SARS-CoV-2 fusion machinery core
- PMID: 35194049
- PMCID: PMC8863989
- DOI: 10.1038/s41467-022-28654-5
Cooperative multivalent receptor binding promotes exposure of the SARS-CoV-2 fusion machinery core
Abstract
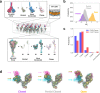
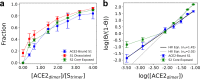
The molecular events that permit the spike glycoprotein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to bind and enter cells are important to understand for both fundamental and therapeutic reasons. Spike proteins consist of S1 and S2 domains, which recognize angiotensin-converting enzyme 2 (ACE2) receptors and contain the viral fusion machinery, respectively. Ostensibly, the binding of spike trimers to ACE2 receptors promotes dissociation of the S1 domains and exposure of the fusion machinery, although the molecular details of this process have yet to be observed. We report the development of bottom-up coarse-grained (CG) models consistent with cryo-electron tomography data, and the use of CG molecular dynamics simulations to investigate viral binding and S2 core exposure. We show that spike trimers cooperatively bind to multiple ACE2 dimers at virion-cell interfaces in a manner distinct from binding between soluble proteins, which processively induces S1 dissociation. We also simulate possible variant behavior using perturbed CG models, and find that ACE2-induced S1 dissociation is primarily sensitive to conformational state populations and the extent of S1/S2 cleavage, rather than ACE2 binding affinity. These simulations reveal an important concerted interaction between spike trimers and ACE2 dimers that primes the virus for membrane fusion and entry.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Update of
-
Cooperative multivalent receptor binding promotes exposure of the SARS-CoV-2 fusion machinery core.bioRxiv [Preprint]. 2021 Jun 7:2021.05.24.445443. doi: 10.1101/2021.05.24.445443. bioRxiv. 2021. Update in: Nat Commun. 2022 Feb 22;13(1):1002. doi: 10.1038/s41467-022-28654-5. PMID: 34127973 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

