Genetic Engineering Systems to Study Human Viral Pathogens from the Coronaviridae Family
- PMID: 35194246
- PMCID: PMC8853348
- DOI: 10.1134/S0026893322010022
Genetic Engineering Systems to Study Human Viral Pathogens from the Coronaviridae Family
Abstract
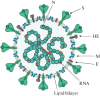
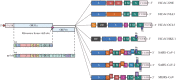
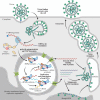
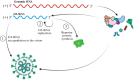
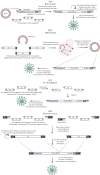
The COVID-19 pandemic caused by the previously unknown SARS-CoV-2 Betacoronavirus made it extremely important to develop simple and safe cellular systems which allow manipulation of the viral genome and high-throughput screening of its potential inhibitors. In this review, we made an attempt at summarizing the currently existing data on genetic engineering systems used to study not only SARS-CoV-2, but also other viruses from the Coronaviridae family. In addition, the review covers the basic knowledge about the structure and the life cycle of coronaviruses.
Keywords: COVID-19; SARS-CoV-2; pseudoviruses; replicons.
© Pleiades Publishing, Inc. 2022, ISSN 0026-8933, Molecular Biology, 2022, Vol. 56, No. 1, pp. 72–89. © Pleiades Publishing, Inc., 2022.Russian Text © The Author(s), 2022, published in Molekulyarnaya Biologiya, 2022, Vol. 56, No. 1, pp. 83–102.
Conflict of interest statement
COMPLIANCE WITH ETHICAL STANDARDSConflict of interest. The authors declare no conflict of interest. Statement on the welfare of humans or animals. This article does not contain any studies involving humans or animals performed by any of the authors.
Figures


Similar articles
-
[Genetic Engineering Systems to Study Human Viral Pathogens from the Coronaviridae Family].Mol Biol (Mosk). 2022 Jan-Feb;56(1):83-102. doi: 10.31857/S0026898422010025. Mol Biol (Mosk). 2022. PMID: 35082260 Review. Russian.
-
[Source of the COVID-19 pandemic: ecology and genetics of coronaviruses (Betacoronavirus: Coronaviridae) SARS-CoV, SARS-CoV-2 (subgenus Sarbecovirus), and MERS-CoV (subgenus Merbecovirus).].Vopr Virusol. 2020;65(2):62-70. doi: 10.36233/0507-4088-2020-65-2-62-70. Vopr Virusol. 2020. PMID: 32515561 Review. Russian.
-
[Etiology of epidemic outbreaks COVID-19 on Wuhan, Hubei province, Chinese People Republic associated with 2019-nCoV (Nidovirales, Coronaviridae, Coronavirinae, Betacoronavirus, Subgenus Sarbecovirus): lessons of SARS-CoV outbreak.].Vopr Virusol. 2020;65(1):6-15. doi: 10.36233/0507-4088-2020-65-1-6-15. Vopr Virusol. 2020. PMID: 32496715 Review. Russian.
-
Comparative analysis of Coronaviridae nucleocapsid and surface glycoprotein sequences.Front Biosci (Landmark Ed). 2020 Jun 1;25(10):1894-1900. doi: 10.2741/4883. Front Biosci (Landmark Ed). 2020. PMID: 32472763
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
Cited by
-
Future trajectory of SARS-CoV-2: Constant spillover back and forth between humans and animals.Virus Res. 2023 Apr 15;328:199075. doi: 10.1016/j.virusres.2023.199075. Epub 2023 Feb 28. Virus Res. 2023. PMID: 36805410 Free PMC article. Review.
-
Expansion of Single Chains Released from a Spherical Cavity.Polymers (Basel). 2022 Dec 30;15(1):198. doi: 10.3390/polym15010198. Polymers (Basel). 2022. PMID: 36616547 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
