One Health compartmental analysis of ESBL-producing Escherichia coli on Reunion Island reveals partitioning between humans and livestock
- PMID: 35194647
- PMCID: PMC9047676
- DOI: 10.1093/jac/dkac054
One Health compartmental analysis of ESBL-producing Escherichia coli on Reunion Island reveals partitioning between humans and livestock
Abstract
Background: Extended-spectrum β-lactamase-producing Escherichia coli (ESBL-Ec) is a major cause of infections worldwide. An understanding of the reservoirs and modes of transmission of these pathogens is essential, to tackle their increasing frequency.
Objectives: We investigated the contributions of various compartments (humans, animals, environment), to human colonization or infection with ESBL-Ec over a 3 year period, on an island.
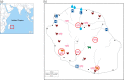
Methods: The study was performed on Reunion Island (Southwest Indian Ocean). We collected ESBL-Ec isolates prospectively from humans, wastewater and livestock between April 2015 and December 2018. Human specimens were recovered from a regional surveillance system representative of the island's health facilities. These isolates were compared with those from livestock and urban/rural wastewater, by whole-genome sequencing.
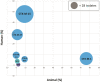
Results: We collected 410 ESBL-Ec isolates: 161 from humans, 161 from wastewater and 88 from animals. Phylogenomic analysis demonstrated high diversity (100 STs), with different STs predominating among isolates from humans (ST131, ST38, ST10) and animals (ST57, ST156). The large majority (90%) of the STs, including ST131, were principally associated with a single compartment. The CTX-M-15, CTX-M-27 and CTX-M-14 enzymes were most common in humans/human wastewater, whereas CTX-M-1 predominated in animals. Isolates of human and animal origin had different plasmids carrying blaCTX-M genes, with the exception of a conserved IncI1-ST3 blaCTX-M-1 plasmid.
Conclusions: These molecular data suggest that, despite their high level of contamination, animals are not a major source of the ESBL-Ec found in humans living on this densely populated high-income island. Public health policies should therefore focus primarily on human-to-human transmission, to prevent human infections with ESBL-Ec.
© The Author(s) 2022. Published by Oxford University Press on behalf of British Society for Antimicrobial Chemotherapy.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

