Hydrop enables droplet-based single-cell ATAC-seq and single-cell RNA-seq using dissolvable hydrogel beads
- PMID: 35195064
- PMCID: PMC8993220
- DOI: 10.7554/eLife.73971
Hydrop enables droplet-based single-cell ATAC-seq and single-cell RNA-seq using dissolvable hydrogel beads
Abstract
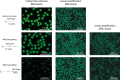
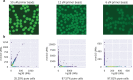
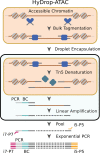
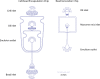
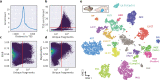
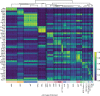
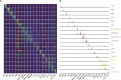
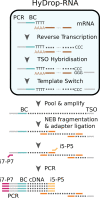
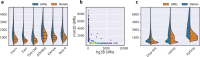
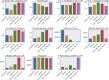
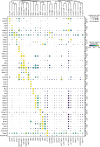
Single-cell RNA-seq and single-cell assay for transposase-accessible chromatin (ATAC-seq) technologies are used extensively to create cell type atlases for a wide range of organisms, tissues, and disease processes. To increase the scale of these atlases, lower the cost and pave the way for more specialized multiome assays, custom droplet microfluidics may provide solutions complementary to commercial setups. We developed HyDrop, a flexible and open-source droplet microfluidic platform encompassing three protocols. The first protocol involves creating dissolvable hydrogel beads with custom oligos that can be released in the droplets. In the second protocol, we demonstrate the use of these beads for HyDrop-ATAC, a low-cost noncommercial scATAC-seq protocol in droplets. After validating HyDrop-ATAC, we applied it to flash-frozen mouse cortex and generated 7996 high-quality single-cell chromatin accessibility profiles in a single run. In the third protocol, we adapt both the reaction chemistry and the capture sequence of the barcoded hydrogel bead to capture mRNA, and demonstrate a significant improvement in throughput and sensitivity compared to previous open-source droplet-based scRNA-seq assays (Drop-seq and inDrop). Similarly, we applied HyDrop-RNA to flash-frozen mouse cortex and generated 9508 single-cell transcriptomes closely matching reference single-cell gene expression data. Finally, we leveraged HyDrop-RNA's high capture rate to analyze a small population of fluorescence-activated cell sorted neurons from the Drosophila brain, confirming the protocol's applicability to low input samples and small cells. HyDrop is currently capable of generating single-cell data in high throughput and at a reduced cost compared to commercial methods, and we envision that HyDrop can be further developed to be compatible with novel (multi) omics protocols.
Keywords: chromosomes; droplet microfluidics; drosophila melanogaster; gene expression; genetics; genomics; human; hydrop; mouse; scATAC-seq; scRNA-seq; single-cell ATAC sequencing; single-cell RNA sequencing.
Plain language summary
Scientists are now able to determine the order of chemical blocks, or nucleic acids, that make up the genetic code. These sequencing tools can be used to identify which genes are active within a biological sample. They do this by extracting and analysing open chromatin (regions of DNA that are accessible to the cell’s machinery), or sequences of RNA (the molecular templates cells use to translate genes into working proteins). Initially, most sequencing tools could only provide an ‘averaged-out’ profile of the genes activated in bulk pieces of tissue which contain multiple types of cell. However, advances in technology have led to new methods that can extract and analyse open chromatin or RNA from individual cells. First, the cells are separated, via a technique called microfluidics, into tiny droplets of water along with a single bead that carries a unique barcode. The cell is then broken apart inside the droplet and the barcode within the bead gets released and attaches itself to the genetic material extracted from the cell. All the genetic material inside the droplets is then pooled together and sequenced. Researchers then use the barcode tags to identify which bits of RNA or DNA belong to each cell. Single-cell sequencing has many advantages, including being able to pinpoint precise genetic differences between healthy and abnormal cells, and to create cell atlases of whole organisms, tissues and microbial communities. But existing methods for extracting chromatin are very expensive, and there were no openly available tools for processing thousands of cells at speed. Furthermore, while several single-cell RNA sequencing tools are already freely available, they are not very sensitive or practical to use. Here, De Rop et al. have developed a new open-source platform called HyDrop that overcomes these barriers. The method entails a new type of barcoded bead and optimised elements of existing microfluidics protocols using open-source reagents. These changes created a more user-friendly workflow and increased sensitivity of sequencing at no additional cost. De Rop et al. used their new platform to screen the RNA and open chromatin of thousands of individuals cells from the brains of mice and flies. HyDrop outperformed other open-source methods when working in RNA-sequencing mode. It also provides the first open-source tool for sequencing open chromatin in single cells. Further improvements are expected as researchers tweak the platform, which for now provides an affordable alternative to existing methods.
© 2022, De Rop et al.
Conflict of interest statement
FD, JI, CB, GH, CF, JJ, KT, VC, JW, GM, Jd, SP, SA No competing interests declared
Figures












Update of
- doi: 10.1101/2021.06.04.447104
Similar articles
-
Efficient chromatin accessibility mapping in situ by nucleosome-tethered tagmentation.Elife. 2020 Nov 16;9:e63274. doi: 10.7554/eLife.63274. Elife. 2020. PMID: 33191916 Free PMC article.
-
Optimized assay for transposase-accessible chromatin by sequencing (ATAC-seq) library preparation from adult Drosophila melanogaster neurons.Sci Rep. 2022 Apr 11;12(1):6043. doi: 10.1038/s41598-022-09869-4. Sci Rep. 2022. PMID: 35411004 Free PMC article.
-
scifi-ATAC-seq: massive-scale single-cell chromatin accessibility sequencing using combinatorial fluidic indexing.Genome Biol. 2024 Apr 8;25(1):90. doi: 10.1186/s13059-024-03235-5. Genome Biol. 2024. PMID: 38589969 Free PMC article.
-
Chromatin accessibility profiling by ATAC-seq.Nat Protoc. 2022 Jun;17(6):1518-1552. doi: 10.1038/s41596-022-00692-9. Epub 2022 Apr 27. Nat Protoc. 2022. PMID: 35478247 Free PMC article. Review.
-
[Advances in assay for transposase-accessible chromatin with high-throughput sequencing].Yi Chuan. 2020 Apr 20;42(4):333-346. doi: 10.16288/j.yczz.19-279. Yi Chuan. 2020. PMID: 32312702 Review. Chinese.
Cited by
-
Biophysical modeling with variational autoencoders for bimodal, single-cell RNA sequencing data.bioRxiv [Preprint]. 2023 May 2:2023.01.13.523995. doi: 10.1101/2023.01.13.523995. bioRxiv. 2023. Update in: Nat Methods. 2024 Aug;21(8):1466-1469. doi: 10.1038/s41592-024-02365-9. PMID: 36712140 Free PMC article. Updated. Preprint.
-
Automatic quality control of single-cell and single-nucleus RNA-seq using valiDrops.NAR Genom Bioinform. 2023 Nov 18;5(4):lqad101. doi: 10.1093/nargab/lqad101. eCollection 2023 Dec. NAR Genom Bioinform. 2023. PMID: 38025048 Free PMC article.
-
Barcodes based on nucleic acid sequences: Applications and challenges (Review).Mol Med Rep. 2025 Jul;32(1):187. doi: 10.3892/mmr.2025.13552. Epub 2025 May 2. Mol Med Rep. 2025. PMID: 40314098 Free PMC article. Review.
-
Biophysical modeling with variational autoencoders for bimodal, single-cell RNA sequencing data.Nat Methods. 2024 Aug;21(8):1466-1469. doi: 10.1038/s41592-024-02365-9. Epub 2024 Jul 25. Nat Methods. 2024. PMID: 39054391
-
The Analysis of the Human Megakaryocyte and Platelet Coding Transcriptome in Healthy and Diseased Subjects.Int J Mol Sci. 2022 Jul 11;23(14):7647. doi: 10.3390/ijms23147647. Int J Mol Sci. 2022. PMID: 35886993 Free PMC article. Review.
References
-
- Bradski G. Automated Calibration of RF On-Wafer Probing and Evaluation of Probe Misalignment Effects Using a Desktop Micro-Factory. Journal of Computer and Communications. 2015;4:122–125. doi: 10.4236/jcc.2016.43009. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

