Identification of electroporation sites in the complex lipid organization of the plasma membrane
- PMID: 35195069
- PMCID: PMC8912918
- DOI: 10.7554/eLife.74773
Identification of electroporation sites in the complex lipid organization of the plasma membrane
Abstract
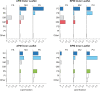
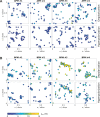
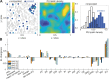
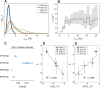
The plasma membrane of a biological cell is a complex assembly of lipids and membrane proteins, which tightly regulate transmembrane transport. When a cell is exposed to strong electric field, the membrane integrity becomes transiently disrupted by formation of transmembrane pores. This phenomenon termed electroporation is already utilized in many rapidly developing applications in medicine including gene therapy, cancer treatment, and treatment of cardiac arrhythmias. However, the molecular mechanisms of electroporation are not yet sufficiently well understood; in particular, it is unclear where exactly pores form in the complex organization of the plasma membrane. In this study, we combine coarse-grained molecular dynamics simulations, machine learning methods, and Bayesian survival analysis to identify how formation of pores depends on the local lipid organization. We show that pores do not form homogeneously across the membrane, but colocalize with domains that have specific features, the most important being high density of polyunsaturated lipids. We further show that knowing the lipid organization is sufficient to reliably predict poration sites with machine learning. Additionally, by analysing poration kinetics with Bayesian survival analysis we show that poration does not depend solely on local lipid arrangement, but also on membrane mechanical properties and the polarity of the electric field. Finally, we discuss how the combination of atomistic and coarse-grained molecular dynamics simulations, machine learning methods, and Bayesian survival analysis can guide the design of future experiments and help us to develop an accurate description of plasma membrane electroporation on the whole-cell level. Achieving this will allow us to shift the optimization of electroporation applications from blind trial-and-error approaches to mechanistic-driven design.
Keywords: electroporation; glycolipids/gangliosides; machine learning; membrane structure; molecular biophysics; molecular dynamics simulations; none; phospholipids; structural biology.
© 2022, Rems et al.
Conflict of interest statement
LR, XT, FZ, SP No competing interests declared, IT, LD Reviewing editor, eLife
Figures











References
-
- Algazi A, Bhatia S, Agarwala S, Molina M, Lewis K, Faries M, Fong L, Levine LP, Franco M, Oglesby A, Ballesteros-Merino C, Bifulco CB, Fox BA, Bannavong D, Talia R, Browning E, Le MH, Pierce RH, Gargosky S, Tsai KK, Twitty C, Daud AI. Intratumoral delivery of tavokinogene telseplasmid yields systemic immune responses in metastatic melanoma patients. Annals of Oncology. 2020;31:532–540. doi: 10.1016/j.annonc.2019.12.008. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

