Discovery and Application of Postnatal Nucleus Pulposus Progenitors Essential for Intervertebral Disc Homeostasis and Degeneration
- PMID: 35195356
- PMCID: PMC9069184
- DOI: 10.1002/advs.202104888
Discovery and Application of Postnatal Nucleus Pulposus Progenitors Essential for Intervertebral Disc Homeostasis and Degeneration
Abstract
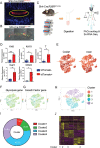
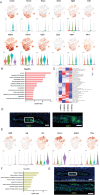
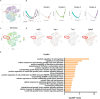
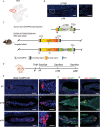
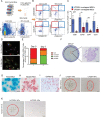
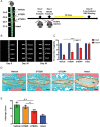
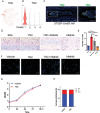
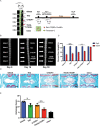
Intervertebral disc degeneration (IDD) results from the dysfunction of nucleus pulposus (NP) cells and the exhaustion of NP progenitors (ProNPs). The cellular applications of NP cells during IDD are currently limited due to the lack of in vivo studies showing whether NP cells are heterogeneous and contain ProNPs throughout postnatal stages. In this study, single-cell RNA sequencing of purified NP cells is used to map four molecularly defined populations and urotensin II receptor (UTS2R)-expressing postnatal ProNPs is identified, which are markedly exhausted during IDD, in mouse and human specimens. The lineage tracing shows that UTS2R+ ProNPs preferentially resides in the NP periphery with its niche factor tenascin-C and give rise to functional NP cells. It is also demonstrated that transplanting UTS2R+ ProNPs with tenascin-C into injured intervertebral discs attenuate the progression of IDD. The study provides a novel NP cell atlas, identified resident ProNPs with regenerative potential, and revealed promising diagnostic and therapeutic targets for IDD.
Keywords: Sc-RNA seq; intervertebral disc; lineage tracing; nucleus pulposus cell atlas; stem cell therapy.
© 2022 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Driscoll T., Jacklyn G., Orchard J., Passmore E., Vos T., Freedman G., Lim S., Punnett L., Ann. Rheum. Dis. 2014, 73, 975. - PubMed
-
- Hoy D., March L., Brooks P., Blyth F., Woolf A., Bain C., Williams G., Smith E., Vos T., Barendregt J., Murray C., Burstein R., Buchbinder R., Ann. Rheum. Dis. 2014, 73, 968. - PubMed
-
- Kido T., Okuyama K., Chiba M., Sasaki H., Seki N., Kamo K., Miyakoshi N., Shimada Y., J. Orthop. Sci. 2016, 21, 419. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
