Mating-Type Switching in Budding Yeasts, from Flip/Flop Inversion to Cassette Mechanisms
- PMID: 35195440
- PMCID: PMC8941940
- DOI: 10.1128/mmbr.00007-21
Mating-Type Switching in Budding Yeasts, from Flip/Flop Inversion to Cassette Mechanisms
Abstract
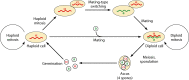
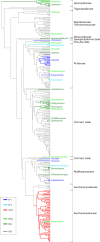
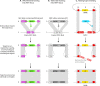
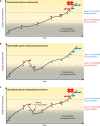
Mating-type switching is a natural but unusual genetic control process that regulates cell identity in ascomycete yeasts. It involves physically replacing one small piece of genomic DNA by another, resulting in replacement of the master regulatory genes in the mating pathway and hence a switch of cell type and mating behavior. In this review, we concentrate on recent progress that has been made on understanding the origins and evolution of mating-type switching systems in budding yeasts (subphylum Saccharomycotina). Because of the unusual nature and the complexity of the mechanism in Saccharomyces cerevisiae, mating-type switching was assumed until recently to have originated only once or twice during yeast evolution. However, comparative genomics analysis now shows that switching mechanisms arose many times independently-at least 11 times in budding yeasts and once in fission yeasts-a dramatic example of convergent evolution. Most of these lineages switch mating types by a flip/flop mechanism that inverts a section of a chromosome and is simpler than the well-characterized 3-locus cassette mechanism (MAT/HML/HMR) used by S. cerevisiae. Mating-type switching (secondary homothallism) is one of the two possible mechanisms by which a yeast species can become self-fertile. The other mechanism (primary homothallism) has also emerged independently in multiple evolutionary lineages of budding yeasts, indicating that homothallism has been favored strongly by natural selection. Recent work shows that HO endonuclease, which makes the double-strand DNA break that initiates switching at the S. cerevisiae MAT locus, evolved from an unusual mobile genetic element that originally targeted a glycolytic gene, FBA1.
Keywords: homothallism; mating type; yeast.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Multiple Reinventions of Mating-type Switching during Budding Yeast Evolution.Curr Biol. 2019 Aug 5;29(15):2555-2562.e8. doi: 10.1016/j.cub.2019.06.056. Epub 2019 Jul 25. Curr Biol. 2019. PMID: 31353182 Free PMC article.
-
A single Ho-induced double-strand break at the MAT locus is lethal in Candida glabrata.PLoS Genet. 2020 Oct 15;16(10):e1008627. doi: 10.1371/journal.pgen.1008627. eCollection 2020 Oct. PLoS Genet. 2020. PMID: 33057400 Free PMC article.
-
The yeast mating-type switching endonuclease HO is a domesticated member of an unorthodox homing genetic element family.Elife. 2020 Apr 27;9:e55336. doi: 10.7554/eLife.55336. Elife. 2020. PMID: 32338594 Free PMC article.
-
An Evolutionary Perspective on Yeast Mating-Type Switching.Genetics. 2017 May;206(1):9-32. doi: 10.1534/genetics.117.202036. Genetics. 2017. PMID: 28476860 Free PMC article. Review.
-
Evolution of Mating in the Saccharomycotina.Annu Rev Microbiol. 2017 Sep 8;71:197-214. doi: 10.1146/annurev-micro-090816-093403. Epub 2017 Jun 28. Annu Rev Microbiol. 2017. PMID: 28657889 Review.
Cited by
-
Yeast sexes: mating types do not determine the sexes in Metschnikowia species.FEMS Yeast Res. 2024 Jan 9;24:foae014. doi: 10.1093/femsyr/foae014. FEMS Yeast Res. 2024. PMID: 38632043 Free PMC article.
-
Fungal sexual reproduction and mating-type loci.Curr Biol. 2025 Jun 9;35(11):R496-R503. doi: 10.1016/j.cub.2025.04.061. Curr Biol. 2025. PMID: 40494303 Free PMC article. Review.
-
A widespread inversion polymorphism conserved among Saccharomyces species is caused by recurrent homogenization of a sporulation gene family.PLoS Genet. 2022 Nov 28;18(11):e1010525. doi: 10.1371/journal.pgen.1010525. eCollection 2022 Nov. PLoS Genet. 2022. PMID: 36441813 Free PMC article.
-
Patterns and mechanisms of fungal genome plasticity.Curr Biol. 2025 Jun 9;35(11):R527-R544. doi: 10.1016/j.cub.2025.04.003. Curr Biol. 2025. PMID: 40494309 Review.
-
Conserved signaling modules regulate filamentous growth in fungi: a model for eukaryotic cell differentiation.Genetics. 2024 Oct 7;228(2):iyae122. doi: 10.1093/genetics/iyae122. Genetics. 2024. PMID: 39239926 Free PMC article. Review.
References
-
- Kurtzman CP, Fell JW, Boekhout T (ed). 2011. The yeasts, a taxonomic study. Elsevier, Amsterdam, The Netherlands.
-
- Phaff HJ, Miller MW, Mrak EM. 1966. The life of yeasts. Harvard University Press, Cambridge, MA.
-
- Zonneveld BJM, Steensma HY. 2003. Mating, sporulation and tetrad analysis in Kluyveromyces lactis, p 151–154. In Wolf K, Breunig K, Barth G (ed), Non-conventional yeasts in genetics, biochemistry and biotechnology. Springer-Verlag, Berlin, Germany.
-
- Lachance MA, Boekhout T, Scorzetti G, Fell JW, Kurtzman CP. 2011. Candida Berkhout (1923), p 987–1278. In Kurtzman CP, Fell JW, Boekhout T (ed), The yeasts, a taxonomic study, vol 2. Elsevier, Amsterdam, The Netherlands.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

