THUMPD1 bi-allelic variants cause loss of tRNA acetylation and a syndromic neurodevelopmental disorder
- PMID: 35196516
- PMCID: PMC9069073
- DOI: 10.1016/j.ajhg.2022.02.001
THUMPD1 bi-allelic variants cause loss of tRNA acetylation and a syndromic neurodevelopmental disorder
Abstract
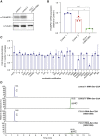
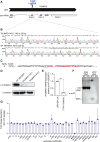
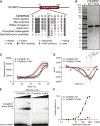
Covalent tRNA modifications play multi-faceted roles in tRNA stability, folding, and recognition, as well as the rate and fidelity of translation, and other cellular processes such as growth, development, and stress responses. Mutations in genes that are known to regulate tRNA modifications lead to a wide array of phenotypes and diseases including numerous cognitive and neurodevelopmental disorders, highlighting the critical role of tRNA modification in human disease. One such gene, THUMPD1, is involved in regulating tRNA N4-acetylcytidine modification (ac4C), and recently was proposed as a candidate gene for autosomal-recessive intellectual disability. Here, we present 13 individuals from 8 families who harbor rare loss-of-function variants in THUMPD1. Common phenotypic findings included global developmental delay, speech delay, moderate to severe intellectual deficiency, behavioral abnormalities such as angry outbursts, facial dysmorphism, and ophthalmological abnormalities. We demonstrate that the bi-allelic variants identified cause loss of function of THUMPD1 and that this defect results in a loss of ac4C modification in small RNAs, and of individually purified tRNA-Ser-CGA. We further corroborate this effect by showing a loss of tRNA acetylation in two CRISPR-Cas9-generated THUMPD1 KO cell lines. In addition, we also show the resultant amino acid substitution that occurs in a missense THUMPD1 allele identified in an individual with compound heterozygous variants results in a marked decrease in THUMPD1 stability and RNA-binding capacity. Taken together, these results suggest that the lack of tRNA acetylation due to THUMPD1 loss of function results in a syndromic form of intellectual disability associated with developmental delay, behavioral abnormalities, hearing loss, and facial dysmorphism.
Keywords: N4-acetylcytidine; NAT10; RNA acetylation; THUMPD1; ac4C; developmental disorder; intellectual disability; tRNA biology; tRNA modifications.
Copyright © 2022 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- El Yacoubi B., Bailly M., de Crécy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012;46:69–95. - PubMed
-
- Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021;22:375–392. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

