Ubiquitin-like 3 as a new protein-sorting factor for small extracellular vesicles
- PMID: 35197392
- PMCID: PMC10511055
- DOI: 10.1247/csf.21078
Ubiquitin-like 3 as a new protein-sorting factor for small extracellular vesicles
Abstract
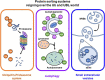
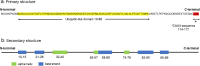
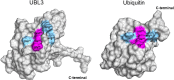
Ubiquitin-like 3 (UBL3) is a well-conserved ubiquitin-like protein (UBL) in eukaryotes and regulates the ubiquitin cascade, but the significant roles of UBL3 in cellular processes remained unknown. Recently, UBL3 was elucidated to be a post-translational modification factor that promotes protein sorting to small extracellular vesicles (sEVs). Proteins sorted into sEVs have been studied as etiologies of sEV-related diseases. Also, there have been attempts to construct drug delivery systems (DDSs) by loading proteins into sEVs. In this review, we introduce the new concept that UBL3 has a critical role in the protein-sorting system and compare structure conservation between UBL3 and other UBLs from an evolutionary perspective. We conclude with future perspectives for the utility of UBL3 in sEV-related diseases and DDS.Key words: UBL3, small extracellular vesicles, protein sorting, ubiquitin-like protein, post-translational modification.
Keywords: UBL3; post-translational modification; protein sorting; small extracellular vesicles; ubiquitin-like protein.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures






References
-
- Ageta, H., Ageta-Ishihara, N., Hitachi, K., Karayel, O., Onouchi, T., Yamaguchi, H., Kahyo, T., Hatanaka, K., Ikegami, K., Yoshioka, Y., Nakamura, K., Kosaka, N., Nakatani, M., Uezumi, A., Ide, T., Tsutsumi, Y., Sugimura, H., Kinoshita, M., Ochiya, T., Mann, M., Setou, M., and Tsuchida, K.. 2018. UBL3 modification influences protein sorting to small extracellular vesicles. Nat. Commun., 9: 3936. - PMC - PubMed
-
- Al-Nedawi, K., Meehan, B., Micallef, J., Lhotak, V., May, L., Guha, A., and Rak, J.. 2008. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol., 10: 619–624. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

