TDP-43 represses cryptic exon inclusion in the FTD-ALS gene UNC13A
- PMID: 35197626
- PMCID: PMC8891019
- DOI: 10.1038/s41586-022-04424-7
TDP-43 represses cryptic exon inclusion in the FTD-ALS gene UNC13A
Abstract
A hallmark pathological feature of the neurodegenerative diseases amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) is the depletion of RNA-binding protein TDP-43 from the nucleus of neurons in the brain and spinal cord1. A major function of TDP-43 is as a repressor of cryptic exon inclusion during RNA splicing2-4. Single nucleotide polymorphisms in UNC13A are among the strongest hits associated with FTD and ALS in human genome-wide association studies5,6, but how those variants increase risk for disease is unknown. Here we show that TDP-43 represses a cryptic exon-splicing event in UNC13A. Loss of TDP-43 from the nucleus in human brain, neuronal cell lines and motor neurons derived from induced pluripotent stem cells resulted in the inclusion of a cryptic exon in UNC13A mRNA and reduced UNC13A protein expression. The top variants associated with FTD or ALS risk in humans are located in the intron harbouring the cryptic exon, and we show that they increase UNC13A cryptic exon splicing in the face of TDP-43 dysfunction. Together, our data provide a direct functional link between one of the strongest genetic risk factors for FTD and ALS (UNC13A genetic variants), and loss of TDP-43 function.
© 2022. The Author(s).
Conflict of interest statement
A.D.G. is a scientific founder of Maze Therapeutics. X.R.M. served as a consultant for Maze Therapeutics. M.P. and L.P. serve as consultants for Target ALS. F.H., B.B.C., D.W.W., K.K., G. Miller, S. Mekhoubad, N.S. and E.G. are employees of Maze Therapeutics, which has filed a patent (63/171,522) on methods to modulate splicing of
Figures









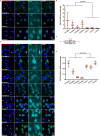

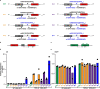
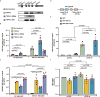

Comment in
-
Mechanism underlying a risk gene in neurodegeneration.Nature. 2022 Mar;603(7899):33-34. doi: 10.1038/d41586-022-00383-1. Nature. 2022. PMID: 35197585 No abstract available.
-
Integrating functional genomics with genetics to understand the biology of ALS and FTD.Med. 2022 Apr 8;3(4):226-227. doi: 10.1016/j.medj.2022.03.008. Med. 2022. PMID: 35590152
-
Cryptic inclusions UNCover losses driving neurodegeneration.Trends Genet. 2022 Sep;38(9):889-891. doi: 10.1016/j.tig.2022.06.004. Epub 2022 Jun 27. Trends Genet. 2022. PMID: 35773026
References
-
- Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. - PubMed
-
- Van Es MA, et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat. Genet. 2009;41:1083–1087. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
- R01 NS104437/NS/NINDS NIH HHS/United States
- R01 AG063780/AG/NIA NIH HHS/United States
- U01 AG057195/AG/NIA NIH HHS/United States
- U19 AG063911/AG/NIA NIH HHS/United States
- P30 AG062677/AG/NIA NIH HHS/United States
- P30 AG062422/AG/NIA NIH HHS/United States
- RF1 NS113636/NS/NINDS NIH HHS/United States
- P01 AG019724/AG/NIA NIH HHS/United States
- U54 NS123743/NS/NINDS NIH HHS/United States
- T32 AG047126/AG/NIA NIH HHS/United States
- RF1 NS120992/NS/NINDS NIH HHS/United States
- R35 NS097263/NS/NINDS NIH HHS/United States
- R35 NS097273/NS/NINDS NIH HHS/United States
- RF1 AG071326/AG/NIA NIH HHS/United States
- T32 GM007231/GM/NIGMS NIH HHS/United States
- T32 HG000044/HG/NHGRI NIH HHS/United States
- P01 NS084974/NS/NINDS NIH HHS/United States
- F32 NS116208/NS/NINDS NIH HHS/United States
- RF1 AG064690/AG/NIA NIH HHS/United States
- U01 AG006786/AG/NIA NIH HHS/United States
- F32 GM139268/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

