Ancient DNA and deep population structure in sub-Saharan African foragers
- PMID: 35197631
- PMCID: PMC8907066
- DOI: 10.1038/s41586-022-04430-9
Ancient DNA and deep population structure in sub-Saharan African foragers
Abstract
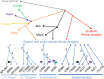
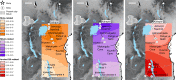
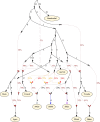
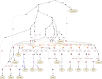
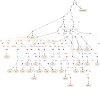
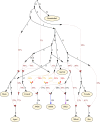
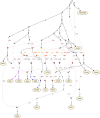
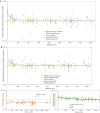
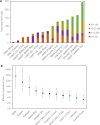
Multiple lines of genetic and archaeological evidence suggest that there were major demographic changes in the terminal Late Pleistocene epoch and early Holocene epoch of sub-Saharan Africa1-4. Inferences about this period are challenging to make because demographic shifts in the past 5,000 years have obscured the structures of more ancient populations3,5. Here we present genome-wide ancient DNA data for six individuals from eastern and south-central Africa spanning the past approximately 18,000 years (doubling the time depth of sub-Saharan African ancient DNA), increase the data quality for 15 previously published ancient individuals and analyse these alongside data from 13 other published ancient individuals. The ancestry of the individuals in our study area can be modelled as a geographically structured mixture of three highly divergent source populations, probably reflecting Pleistocene interactions around 80-20 thousand years ago, including deeply diverged eastern and southern African lineages, plus a previously unappreciated ubiquitous distribution of ancestry that occurs in highest proportion today in central African rainforest hunter-gatherers. Once established, this structure remained highly stable, with limited long-range gene flow. These results provide a new line of genetic evidence in support of hypotheses that have emerged from archaeological analyses but remain contested, suggesting increasing regionalization at the end of the Pleistocene epoch.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

