Evaluation of Bacteriophage Cocktail on Septicemia Caused by Colistin-Resistant Klebsiella pneumoniae in Mice Model
- PMID: 35197852
- PMCID: PMC8860340
- DOI: 10.3389/fphar.2022.778676
Evaluation of Bacteriophage Cocktail on Septicemia Caused by Colistin-Resistant Klebsiella pneumoniae in Mice Model
Abstract
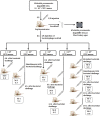
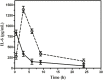
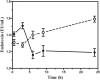
Objective: The emergence of resistance against last-resort antibiotics, carbapenem and colistin, in Klebsiella pneumoniae has been reported across the globe. Bacteriophage therapy seems to be one of the most promising alternatives. This study aimed to optimize the quantity and frequency of bacteriophage cocktail dosage/s required to eradicate the Klebsiella pneumoniae bacteria in immunocompetent septicemic mice. Methods: The three most active phages ɸKpBHU4, ɸKpBHU7, and ɸKpBHU14 characterized by molecular and TEM analyses were in the form of cocktail and was given intraperitoneally to mice after inducing the septicemia mice model with a constant dose of 8 × 107 colony-forming unit/mouse (CFU/mouse) Klebsiella pneumoniae. After that, the efficacy of the phage cocktail was analyzed at different dosages, that is, in increasing, variable, constant, and repeated dosages. Furthermore, interleukin-6 and endotoxin levels were estimated with variable doses of phage cocktail. Results: We have elucidated that phage therapy is effective against the Klebsiella pneumoniae septicemia mice model and is a promising alternative to antibiotic treatments. Our work delineates that a single dose of phage cocktail with 1 × 105 plaque-forming unit/mouse (PFU/mouse) protects the mice from fatal outcomes at any stage of septicemia. However, a higher phage dosage of 1 × 1012 PFU/mice is fatal when given at the early hours of septicemia, while this high dose is not fatal at the later stages of septicemia. Moreover, multiple repeated dosages are required to eradicate the bacteria from peripheral blood. In addition, the IL-6 levels in the 1 × 105 PFU/mouse group remain lower, but in the 1 × 1012 PFU/mouse group remains high at all points, which were associated with fatal outcomes. Conclusion: Our study showed that the optimized relatively lower and multiple dosages of phage cocktails with the strict monitoring of vitals in clinical settings might cure septicemia caused by MDR bacteria with different severity of infection.
Keywords: IL-6; Klebsiella pneumoniae; endotoxin; phage cocktail; septicemia.
Copyright © 2022 Singh, Singh, Rathor, Chaudhry, Singh and Nath.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Adams M. H. (1959). Bacteriophages. New York: Interscience Publishers.
-
- Capone A., Giannella M., Fortini D., Giordano A., Meledandri M., Ballardini M., et al. (2013). High Rate of Colistin Resistance Among Patients with Carbapenem-Resistant Klebsiella pneumoniae Infection Accounts for an Excess of Mortality. Clin. Microbiol. Infect. 19 (1), E23–E30. 10.1111/1469-0691.12070 - DOI - PubMed
-
- Czajkowski R., Ozymko Z., de Jager V., Siwinska J., Smolarska A., Ossowicki A., et al. (2015). Genomic, Proteomic and Morphological Characterization of Two Novel Broad Host Lytic Bacteriophages ΦPD10.3 and ΦPD23.1 Infecting Pectinolytic Pectobacterium Spp. And Dickeya Spp. PLOS ONE 10 (3), e0119812. 10.1371/journal.pone.0119812 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

