Single Nucleotide Polymorphism Detection for Peach Gummosis Disease Resistance by Genome-Wide Association Study
- PMID: 35197988
- PMCID: PMC8858797
- DOI: 10.3389/fpls.2021.763618
Single Nucleotide Polymorphism Detection for Peach Gummosis Disease Resistance by Genome-Wide Association Study
Abstract
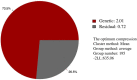
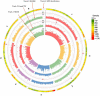
Peach gummosis is one of the most widespread and destructive diseases. It causes growth stunting, yield loss, branch, trunk, and tree death, and is becoming a restrictive factor in healthy and sustainable development of peach production. Although a locus has been identified based on bi-parental quantitative trait locus (QTL) mapping, selection of gummosis-resistant cultivars remains challenging due to the lack of resistant parents and of the complexity of an inducing factor. In this study, an integrated approach of genome-wide association study (GWAS) and comparative transcriptome was used to elucidate the genetic architecture associated with the disease using 195 accessions and 145,456 genome-wide single nucleotide polymorphisms (SNPs). The broad-sense and narrow-sense heritabilities were estimated using 2-year phenotypic data and genotypic data, which gave high values of 70 and 73%, respectively. Evaluation of population structure by neighbor-joining and principal components analysis (PCA) clustered all accessions into three major groups and six subgroups, mainly according to fruit shape, hairy vs. glabrous fruit skin, pedigree, geographic origin, and domestication history. Five SNPs were found to be significantly associated with gummosis disease resistance, of which SNPrs285957, located on chromosome6 across 28 Mb, was detected by both the BLINK and the FarmCPU model. Six candidate genes flanked by or harboring the significant SNPs, previously implicated in biotic stress tolerance, were significantly associated with this resistance. Two highly resistant accessions were identified with low disease severity, which could be potential sources of resistance genes for breeding. Our results provide a fresh insight into the genetic control of peach gummosis disease.
Keywords: QTLs; candidate genes; genome-wide association study; gummosis disease; peach.
Copyright © 2022 Li, Wang, Su, Zhou, Zhang, Du, Zhou, Gan, Jin, Zhang, Cao, Fang, Wang, Jia, Gao and Ye.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Beckman T. G., Reilly C. C. (2005). Relative ausceptibility of ornamental peach cultivars to fungal gummosis (Botryosphaeria dothidea). J. Am. Pomol. Soc. 60 149–154.
-
- Beckman T. G., Reilly C. C., Pusey P. L., Hotchkiss M. (2011). Progress in the management of peach fungal gummosis (Botryosphaeria dothidea) in the southeastern US peach industry. Mater. Sci. Appl. 65 192–200.
LinkOut - more resources
Full Text Sources
Miscellaneous

