Overexpression of S100A9 in obesity impairs macrophage differentiation via TLR4-NFkB-signaling worsening inflammation and wound healing
- PMID: 35198063
- PMCID: PMC8825590
- DOI: 10.7150/thno.67174
Overexpression of S100A9 in obesity impairs macrophage differentiation via TLR4-NFkB-signaling worsening inflammation and wound healing
Abstract
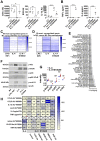
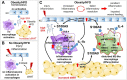
Rationale: In obesity the fine-tuned balance of macrophage phenotypes is disturbed towards a dominance of pro-inflammatory macrophages resulting in exacerbation and persistence of inflammation and impaired tissue repair. However, the underlying mechanisms are still poorly understood. Methods: Impact of obesity on macrophage differentiation was studied in high fat diet induced obese and db/db mice during skin inflammation and wound repair, respectively. Mechanisms of S100A9-mediated effects on macrophage differentiation was studied on in vitro generated macrophages by genomic and proteomic approaches. The role of S100A9 on macrophage differentiation was investigated by pharmacological inhibition of S100A9 during skin inflammation and wound repair in obese and db/db mice. Results: We demonstrate an overexpression of S100A9 in conditions of obesity-associated disturbed macrophage differentiation in the skin. We show that saturated free fatty acids (SFA), which are increased in obesity, together with S100A9 induce TLR4 and inflammasome-dependent IL-1β release in macrophages which in turn amplifies S100A9 expression initiating a vicious cycle of sustained S100A9 overexpression in skin inflammation in obesity. We reveal a yet unrecognized impact of obesity-associated S100A9 overexpression on macrophage differentiation. S100A9 binding to TLR4 and activation of NFkB attenuates development of M2-like macrophages and induces pro-inflammatory functions in these cells. Consequently, inhibition of S100A9 restores disturbed M2-like macrophage differentiation in mouse models of obesity-associated skin inflammation and wound repair. Similarly, breaking the vicious cycle of S100A9 overexpression by dietary reduction of SFA restored M2-like macrophage activation. Improvement of skin inflammation and wound repair upon reduction of S100A9 by pharmacological inhibition or by reduction of SFA uncovers the pathogenic role of S100A9 overexpression in obesity. Conclusion: This study identifies S100A9 as a previously unrecognized vital component in obesity-associated disturbed macrophage differentiation and subsequent impaired regulation of inflammation and wound repair. The findings open new opportunities for therapeutic implications for inflammatory diseases and wound repair in obesity.
Keywords: S100A9; macrophage differentiation; obesity; skin inflammation; wound healing.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

