Enzymatic activity of glycosyltransferase GLT8D1 promotes human glioblastoma cell migration
- PMID: 35198895
- PMCID: PMC8850796
- DOI: 10.1016/j.isci.2022.103842
Enzymatic activity of glycosyltransferase GLT8D1 promotes human glioblastoma cell migration
Abstract
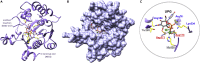
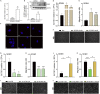
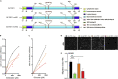
Glioblastoma (GBM) is the most aggressive primary brain tumor characterized by infiltrative growth of malignant glioma cells into the surrounding brain parenchyma. In this study, our analysis of GBM patient cohorts revealed a significantly higher expression of Glycosyltransferase 8 domain containing 1 (GLT8D1) compared to normal brain tissue and could be associated with impaired patient survival. Increased in vitro expression of GLT8D1 significantly enhanced migration of two different sphere-forming GBM cell lines. By in silico analysis we predicted the 3D-structure as well as the active site residues of GLT8D1. The introduction of point mutations in the predicted active site reduced its glycosyltransferase activity in vitro and consequently impaired GBM tumor cell migration. Examination of GLT8D1 interaction partners by LC-MS/MS implied proteins associated with cytoskeleton and intracellular transport as potential substrates. In conclusion, we demonstrated that the enzymatic activity of glycosyltransferase GLT8D1 promotes GBM cell migration.
Keywords: Biochemistry; Cancer; Cell biology; Glycobiology.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Bougnaud S., Golebiewska A., Oudin A., Keunen O., Harter P.N., Mäder L., Azuaje F., Fritah S., Stieber D., Kaoma T., et al. Molecular crosstalk between tumour and brain parenchyma instructs histopathological features in glioblastoma. Oncotarget. 2016;7:31955–31971. doi: 10.18632/oncotarget.7454. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

