Zrsr2 and functional U12-dependent spliceosome are necessary for follicular development
- PMID: 35198906
- PMCID: PMC8850803
- DOI: 10.1016/j.isci.2022.103860
Zrsr2 and functional U12-dependent spliceosome are necessary for follicular development
Abstract
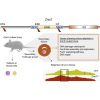
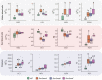
ZRSR2 is a splicing factor involved in recognition of 3'-intron splice sites that is frequently mutated in myeloid malignancies and several tumors; however, the role of mutations of Zrsr2 in other tissues has not been analyzed. To explore the biological role of ZRSR2, we generated three Zrsr2 mutant mouse lines. All Zrsr2 mutant lines exhibited blood cell anomalies, and in two lines, oogenesis was blocked at the secondary follicle stage. RNA-seq of Zrsr2 mu secondary follicles showed aberrations in gene expression and showed altered alternative splicing (AS) events involving enrichment of U12-type intron retention (IR), supporting the functional Zrsr2 action in minor spliceosomes. IR events were preferentially associated with centriole replication, protein phosphorylation, and DNA damage checkpoint. Notably, we found alterations in AS events of 50 meiotic genes. These results indicate that ZRSR2 mutations alter splicing mainly in U12-type introns, which may affect peripheral blood cells, and impede oogenesis and female fertility.
Keywords: Biological sciences; Molecular biology; Transcriptomics.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Comment in
-
Differential alternative splicing analysis links variation in ZRSR2 to a novel type of oral-facial-digital syndrome.Genet Med. 2024 Apr;26(4):101059. doi: 10.1016/j.gim.2023.101059. Epub 2023 Dec 27. Genet Med. 2024. PMID: 38158857
References
-
- Alen F., Gomez-Redondo I., Rivera P., Suarez J., Ramos-Ibeas P., Pericuesta E., Fernandez-Gonzalez R., Perez-Cerezales S., Horiuchi K., Orio L., et al. Sex-dimorphic behavioral alterations and altered neurogenesis in U12 intron splicing-defective Zrsr1 mutant mice. Int. J. Mol. Sci. 2019;20:3543. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

