Exploring Noncovalent Protease Inhibitors for the Treatment of Severe Acute Respiratory Syndrome and Severe Acute Respiratory Syndrome-Like Coronaviruses
- PMID: 35199517
- PMCID: PMC8887654
- DOI: 10.1021/acsinfecdis.1c00631
Exploring Noncovalent Protease Inhibitors for the Treatment of Severe Acute Respiratory Syndrome and Severe Acute Respiratory Syndrome-Like Coronaviruses
Abstract
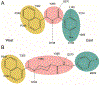
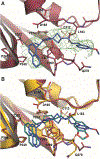
Over the last 20 years, both severe acute respiratory syndrome coronavirus-1 and severe acute respiratory syndrome coronavirus-2 have transmitted from animal hosts to humans causing zoonotic outbreaks of severe disease. Both viruses originate from a group of betacoronaviruses known as subgroup 2b. The emergence of two dangerous human pathogens from this group along with previous studies illustrating the potential of other subgroup 2b members to transmit to humans has underscored the need for antiviral development against them. Coronaviruses modify the host innate immune response in part through the reversal of ubiquitination and ISGylation with their papain-like protease (PLpro). To identify unique or overarching subgroup 2b structural features or enzymatic biases, the PLpro from a subgroup 2b bat coronavirus, BtSCoV-Rf1.2004, was biochemically and structurally evaluated. This evaluation revealed that PLpros from subgroup 2b coronaviruses have narrow substrate specificity for K48 polyubiquitin and ISG15 originating from certain species. The PLpro of BtSCoV-Rf1.2004 was used as a tool alongside PLpro of CoV-1 and CoV-2 to design 30 novel noncovalent drug-like pan subgroup 2b PLpro inhibitors that included determining the effects of using previously unexplored core linkers within these compounds. Two crystal structures of BtSCoV-Rf1.2004 PLpro bound to these inhibitors aided in compound design as well as shared structural features among subgroup 2b proteases. Screening of these three subgroup 2b PLpros against this novel set of inhibitors along with cytotoxicity studies provide new directions for pan-coronavirus subgroup 2b antiviral development of PLpro inhibitors.
Keywords: COVID-19; ISG5; PLpro; coronavirus; severe acute respiratory syndrome 2; ubiquitin.
Conflict of interest statement
Conflict of Interest
BTF, SDP, RAT, and RJH have submitted a provisional application U.S.S.N. 62/992,895 pertaining to the work enclosed in the manuscript. Also, U.S.S.N 63/086,137 has been submitted by BTF, DAA, RSB, IAD, JH, EO, MP, YPS, RAT, BSC, DC, SDP pertaining to work enclosed in the manuscript.
Figures

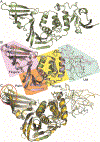
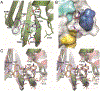


References
-
- Li W; Shi Z; Yu M; Ren W; Smith C; Epstein JH; Wang H; Crameri G; Hu Z; Zhang H; Zhang J; McEachern J; Field H; Daszak P; Eaton BT; Zhang S; Wang LF, Bats are natural reservoirs of SARS-like coronaviruses. Science 2005, 310 (5748), 676–9. - PubMed
-
- Menachery VD; Yount BL Jr.; Debbink K; Agnihothram S; Gralinski LE; Plante JA; Graham RL; Scobey T; Ge XY; Donaldson EF; Randell SH; Lanzavecchia A; Marasco WA; Shi ZL; Baric RS, A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med 2015, 21 (12), 1508–13. - PMC - PubMed
-
- Lam TT; Jia N; Zhang YW; Shum MH; Jiang JF; Zhu HC; Tong YG; Shi YX; Ni XB; Liao YS; Li WJ; Jiang BG; Wei W; Yuan TT; Zheng K; Cui XM; Li J; Pei GQ; Qiang X; Cheung WY; Li LF; Sun FF; Qin S; Huang JC; Leung GM; Holmes EC; Hu YL; Guan Y; Cao WC, Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020, 583 (7815), 282–285. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

