Conplastic strains for identification of retrograde effects of mitochondrial dna variation on cardiometabolic traits in the spontaneously hypertensive rat
- PMID: 35199537
- PMCID: PMC9054184
- DOI: 10.33549/physiolres.934740
Conplastic strains for identification of retrograde effects of mitochondrial dna variation on cardiometabolic traits in the spontaneously hypertensive rat
Abstract
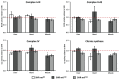
Mitochondrial retrograde signaling is a pathway of communication from mitochondria to the nucleus. Recently, natural mitochondrial genome (mtDNA) polymorphisms (haplogroups) received increasing attention in the pathophysiology of human common diseases. However, retrograde effects of mtDNA variants on such traits are difficult to study in humans. The conplastic strains represent key animal models to elucidate regulatory roles of mtDNA haplogroups on defined nuclear genome background. To analyze the relationship between mtDNA variants and cardiometabolic traits, we derived a set of rat conplastic strains (SHR-mtBN, SHR-mtF344 and SHR-mtLEW), harboring all major mtDNA haplotypes present in common inbred strains on the nuclear background of the spontaneously hypertensive rat (SHR). The BN, F344 and LEW mtDNA differ from the SHR in multiple amino acid substitutions in protein coding genes and also in variants of tRNA and rRNA genes. Different mtDNA haplotypes were found to predispose to various sets of cardiometabolic phenotypes which provided evidence for significant retrograde effects of mtDNA in the SHR. In the future, these animals could be used to decipher individual biochemical components involved in the retrograde signaling.
Conflict of interest statement
There is no conflict of interest.
Figures


References
-
- AITMAN TJ, GLAZIER AM, WALLACE CA, COOPER LD, NORSWORTHY PJ, WAHID FN, AL-MAJALI KM, TREMBLING PM, MANN CJ, SHOULDERS CC, GRAF D, ST LEZIN E, KURTZ TW, KREN V, PRAVENEC M, IBRAHIMI A, ABUMRAD NA, STANTON LW, SCOTT J. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat Genet. 1999;21:76–83. doi: 10.1038/5013. - DOI - PubMed
-
- AW WC, TOWARNICKI SG, MELVIN RG, YOUNGSON NA, GARVIN MR, HU Y, NIELSEN S, THOMAS T, PICKFORD R, BUSTAMANTE S, VILA-SANJURJO A, SMYTH GK, BALLARD JWO. Genotype to phenotype: Diet-by-mitochondrial DNA haplotype interactions drive metabolic flexibility and organismal fitness. PLoS Genet. 2018;14:e1007735. doi: 10.1371/journal.pgen.1007735. - DOI - PMC - PubMed
-
- CYPROVÁ M, BENÁK D, HLAVÁČKOVÁ M, ŠILHAVÝ J, PRAVENEC M, KOLÁŘ F, NECKÁŘ J. Cardiac ischemic tolerance of spontaneously hypertensive rats with replaced mitochondrial DNA: Effect of adaptation to chronic hypoxia. J Mol Cell Cardiol. 2018;120(Suppl 1):5. doi: 10.1016/j.yjmcc.2018.05.027. - DOI
-
- DUNHAM-SNARY KJ, SANDEL MW, SAMMY MJ, WESTBROOK DG, XIAO R, MCMONIGLE RJ, RATCLIFFE WF, PENN A, YOUNG ME, BALLINGER SW. Mitochondrial - nuclear genetic interaction modulates whole body metabolism, adiposity and gene expression in vivo. EBioMedicine. 2018;36:316–328. doi: 10.1016/j.ebiom.2018.08.036. - DOI - PMC - PubMed
