A revised view on the evolution of glutamine synthetase isoenzymes in plants
- PMID: 35199893
- PMCID: PMC9310647
- DOI: 10.1111/tpj.15712
A revised view on the evolution of glutamine synthetase isoenzymes in plants
Abstract
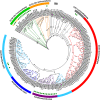
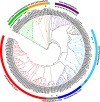
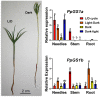
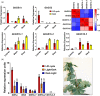
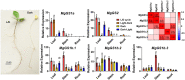
Glutamine synthetase (GS) is a key enzyme responsible for the incorporation of inorganic nitrogen in the form of ammonium into the amino acid glutamine. In plants, two groups of functional GS enzymes are found: eubacterial GSIIb (GLN2) and eukaryotic GSIIe (GLN1/GS). Only GLN1/GS genes are found in vascular plants, which suggests that they are involved in the final adaptation of plants to terrestrial life. The present phylogenetic study reclassifies the different GS genes of seed plants into three clusters: GS1a, GS1b and GS2. The presence of genes encoding GS2 has been expanded to Cycadopsida gymnosperms, which suggests the origin of this gene in a common ancestor of Cycadopsida, Ginkgoopsida and angiosperms. GS1a genes have been identified in all gymnosperms, basal angiosperms and some Magnoliidae species. Previous studies in conifers and the gene expression profiles obtained in ginkgo and magnolia in the present work could explain the absence of GS1a in more recent angiosperm species (e.g. monocots and eudicots) as a result of the redundant roles of GS1a and GS2 in photosynthetic cells. Altogether, the results provide a better understanding of the evolution of plant GS isoenzymes and their physiological roles, which is valuable for improving crop nitrogen use efficiency and productivity. This new view of GS evolution in plants, including a new cytosolic GS group (GS1a), has important functional implications in the context of plant metabolism adaptation to global changes.
Keywords: adaptation; glutamine synthetase; new gene classification; nitrogen metabolism; phylogeny; plant evolution.
© 2022 The Authors. The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflicts of interest associated with this work.
Figures

References
-
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. & Lipman, D.J. (1990) Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. - PubMed
-
- Ávila, C. , Suárez, M.F. , Gómez‐Maldonado, J. & Cánovas, F.M. (2001) Spatial and temporal expression of two cytosolic glutamine synthetase genes in scots pine: functional implications on nitrogen metabolism during early stages of conifer development. The Plant Journal, 25, 93–102. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

